Optimizing the evaluation of gene-targeted panels for tumor mutational burden estimation
- PMID: 34702927
- PMCID: PMC8548549
- DOI: 10.1038/s41598-021-00626-7
Optimizing the evaluation of gene-targeted panels for tumor mutational burden estimation
Abstract
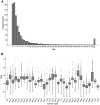
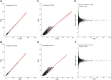
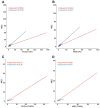
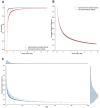
Though whole exome sequencing (WES) is the gold-standard for measuring tumor mutational burden (TMB), the development of gene-targeted panels enables cost-effective TMB estimation. With the growing number of panels in clinical trials, developing a statistical method to effectively evaluate and compare the performance of different panels is necessary. The mainstream method uses R-squared value to measure the correlation between the panel-based TMB and WES-based TMB. However, the performance of a panel is usually overestimated via R-squared value based on the long-tailed TMB distribution of the dataset. Herein, we propose angular distance, a measurement used to compute the extent of the estimated bias. Our extensive in silico analysis indicates that the R-squared value reaches a plateau after the panel size reaches 0.5 Mb, which does not adequately characterize the performance of the panels. In contrast, the angular distance is still sensitive to the changes in panel sizes when the panel size reaches 6 Mb. In particular, R-squared values between the hypermutation-included dataset and the non-hypermutation dataset differ widely across many cancer types, whereas the angular distances are highly consistent. Therefore, the angular distance is more objective and logical than R-squared value for evaluating the accuracy of TMB estimation for gene-targeted panels.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

