Evaluation of a custom QIAseq targeted DNA panel with 164 ancestry informative markers sequenced with the Illumina MiSeq
- PMID: 34702940
- PMCID: PMC8548529
- DOI: 10.1038/s41598-021-99933-2
Evaluation of a custom QIAseq targeted DNA panel with 164 ancestry informative markers sequenced with the Illumina MiSeq
Abstract
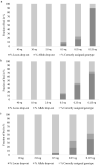
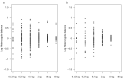
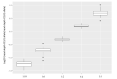
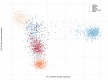
Introduction of new methods requires meticulous evaluation before they can be applied to forensic genetic case work. Here, a custom QIAseq Targeted DNA panel with 164 ancestry informative markers was assessed using the MiSeq sequencing platform. Concordance, sensitivity, and the capability for analysis of mixtures were tested. The assay gave reproducible and nearly concordant results with an input of 10 and 2 ng DNA. Lower DNA input led to an increase in both locus and allele drop-outs, and a higher variation in heterozygote balance. Locus or allele drop-outs in the samples with less than 2 ng DNA input were not necessarily associated with the overall performance of a locus. Thus, the QIAseq assay will be difficult to implement in a forensic genetic setting where the sample material is often scarce and of poor quality. With equal or near equal mixture ratios, the mixture DNA profiles were easily identified by an increased number of imbalanced heterozygotes. For more skewed mixture ratios, the mixture DNA profiles were identified by an increased noise level. Lastly, individuals from Great Britain and the Middle East were investigated. The Middle Eastern individuals showed a greater affinity with South European populations compared to North European populations.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Evaluation of a custom GeneRead™ massively parallel sequencing assay with 210 ancestry informative SNPs using the Ion S5™ and MiSeq platforms.Forensic Sci Int Genet. 2021 Jan;50:102411. doi: 10.1016/j.fsigen.2020.102411. Epub 2020 Nov 2. Forensic Sci Int Genet. 2021. PMID: 33176271
-
Massively parallel sequencing of forensic STRs and SNPs using the Illumina® ForenSeq™ DNA Signature Prep Kit on the MiSeq FGx™ Forensic Genomics System.Forensic Sci Int Genet. 2017 Nov;31:135-148. doi: 10.1016/j.fsigen.2017.09.003. Epub 2017 Sep 8. Forensic Sci Int Genet. 2017. PMID: 28938154
-
Evaluation of the Illumina(®) Beta Version ForenSeq™ DNA Signature Prep Kit for use in genetic profiling.Forensic Sci Int Genet. 2016 Jan;20:20-29. doi: 10.1016/j.fsigen.2015.09.009. Epub 2015 Sep 21. Forensic Sci Int Genet. 2016. PMID: 26433485
-
Evaluation of microhaplotypes in forensic kinship analysis from a Swedish population perspective.Int J Legal Med. 2021 Jul;135(4):1151-1160. doi: 10.1007/s00414-021-02509-y. Epub 2021 Jan 28. Int J Legal Med. 2021. PMID: 33506298 Free PMC article.
-
The analysis of ancestry with small-scale forensic panels of genetic markers.Emerg Top Life Sci. 2021 Sep 24;5(3):443-453. doi: 10.1042/ETLS20200327. Emerg Top Life Sci. 2021. PMID: 33949669 Review.
Cited by
-
Applying Unique Molecular Indices with an Extensive All-in-One Forensic SNP Panel for Improved Genotype Accuracy and Sensitivity.Genes (Basel). 2023 Mar 29;14(4):818. doi: 10.3390/genes14040818. Genes (Basel). 2023. PMID: 37107576 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

