Control of APOBEC3B induction and cccDNA decay by NF-κB and miR-138-5p
- PMID: 34704004
- PMCID: PMC8523871
- DOI: 10.1016/j.jhepr.2021.100354
Control of APOBEC3B induction and cccDNA decay by NF-κB and miR-138-5p
Abstract
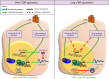
Background & aims: Immune-mediated induction of cytidine deaminase APOBEC3B (A3B) expression leads to HBV covalently closed circular DNA (cccDNA) decay. Here, we aimed to decipher the signalling pathway(s) and regulatory mechanism(s) involved in A3B induction and related HBV control.
Methods: Differentiated HepaRG cells (dHepaRG) knocked-down for NF-κB signalling components, transfected with siRNA or micro RNAs (miRNA), and primary human hepatocytes ± HBV or HBVΔX or HBV-RFP, were treated with lymphotoxin beta receptor (LTβR)-agonist (BS1). The biological outcomes were analysed by reverse transcriptase-qPCR, immunoblotting, luciferase activity, chromatin immune precipitation, electrophoretic mobility-shift assay, targeted-bisulfite-, miRNA-, RNA-, genome-sequencing, and mass-spectrometry.
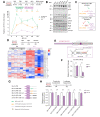
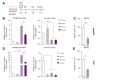
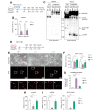
Results: We found that canonical and non-canonical NF-κB signalling pathways are mandatory for A3B induction and anti-HBV effects. The degree of immune-mediated A3B production is independent of A3B promoter demethylation but is controlled post-transcriptionally by the miRNA 138-5p expression (hsa-miR-138-5p), promoting A3B mRNA decay. Hsa-miR-138-5p over-expression reduced A3B levels and its antiviral effects. Of note, established infection inhibited BS1-induced A3B expression through epigenetic modulation of A3B promoter. Twelve days of treatment with a LTβR-specific agonist BS1 is sufficient to reduce the cccDNA pool by 80% without inducing significant damages to a subset of cancer-related host genes. Interestingly, the A3B-mediated effect on HBV is independent of the transcriptional activity of cccDNA as well as on rcDNA synthesis.
Conclusions: Altogether, A3B represents the only described enzyme to target both transcriptionally active and inactive cccDNA. Thus, inhibiting hsa-miR-138-5p expression should be considered in the combinatorial design of new therapies against HBV, especially in the context of immune-mediated A3B induction.
Lay summary: Immune-mediated induction of cytidine deaminase APOBEC3B is transcriptionally regulated by NF-κB signalling and post-transcriptionally downregulated by hsa-miR-138-5p expression, leading to cccDNA decay. Timely controlled APOBEC3B-mediated cccDNA decay occurs independently of cccDNA transcriptional activity and without damage to a subset of cancer-related genes. Thus, APOBEC3B-mediated cccDNA decay could offer an efficient therapeutic alternative to target hepatitis B virus chronic infection.
Keywords: A20, tumour necrosis factor alpha-induced protein 3; APOBEC3A/A3A, apolipoprotein B mRNA editing catalytic polypeptide-like A; APOBEC3B; APOBEC3B/A3B, apolipoprotein B mRNA editing catalytic polypeptide-like B; APOBEC3G/A3G, apolipoprotein B mRNA editing catalytic polypeptide-like G; BCA, bicinchoninic acid assay; CHB, chronic hepatitis B; CXCL10, C-X-C motif chemokine ligand 10; ChIP, chromatin immune precipitation; EMSA, electrophoretic mobility-shift assay; H3K4Me3, histone 3 lysine 4 trimethylation; HBx; Hepatitis B virus; IFNα/γ, interferon alpha/gamma; IKKα/β, IκB kinase alpha/beta; JMJD8, jumonji domain containing 8; LPS, lipopolysaccharide; LTβR, lymphotoxin beta receptor; MAPK, mitogen-activated protein kinase; NEMO, NF-κB essential modulator; NF-κB; NF-κB, nuclear factor kappa B; NIK, NF-κB inducing kinase; NT, non-treated; RT-qPCR, reverse transcription-quantitative PCR; RelA, NF-κB p65 subunit; TNF, tumour necrosis factor; UBE2V1, ubiquitin conjugating enzyme E2 V1; UTR, untranslated region; cccDNA; cccDNA, covalently closed circular DNA; d.p.i., days post infection; miRNA; miRNA, micro RNA; siCTRL, siRNA control.
© 2021 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

