Establishment of a Publicly Available Core Genome Multilocus Sequence Typing Scheme for Clostridium perfringens
- PMID: 34704797
- PMCID: PMC8549748
- DOI: 10.1128/Spectrum.00533-21
Establishment of a Publicly Available Core Genome Multilocus Sequence Typing Scheme for Clostridium perfringens
Abstract
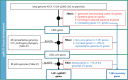
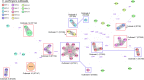
Clostridium perfringens is a spore-forming anaerobic pathogen responsible for a variety of histotoxic and intestinal infections in humans and animals. High-resolution genotyping aiming to identify bacteria at strain level has become increasingly important in modern microbiology to understand pathogen transmission pathways and to tackle infection sources. This study aimed at establishing a publicly available genome-wide multilocus sequence-typing (MLST) scheme for C. perfringens. A total of 1,431 highly conserved core genes (1.34 megabases; 50% of the reference genome genes) were indexed for a core genome-based MLST (cgMLST) scheme for C. perfringens. The scheme was applied to 282 ecologically and geographically diverse genomes, showing that the genotyping results of cgMLST were highly congruent with the core genome-based single-nucleotide-polymorphism typing in terms of resolution and tree topology. In addition, the cgMLST provided a greater discrimination than classical MLST methods for C. perfringens. The usability of the scheme for outbreak analysis was confirmed by reinvestigating published outbreaks of C. perfringens-associated infections in the United States and the United Kingdom. In summary, a publicly available scheme and an allele nomenclature database for genomic typing of C. perfringens have been established and can be used for broad-based and standardized epidemiological studies. IMPORTANCE Global epidemiological surveillance of bacterial pathogens is enhanced by the availability of standard tools and sharing of typing data. The use of whole-genome sequencing has opened the possibility for high-resolution characterization of bacterial strains down to the clonal and subclonal levels. Core genome multilocus sequence typing is a robust system that uses highly conserved core genes for deep genotyping. The method has been successfully and widely used to describe the epidemiology of various bacterial species. Nevertheless, a cgMLST typing scheme for Clostridium perfringens is currently not publicly available. In this study, we (i) developed a cgMLST typing scheme for C. perfringens, (ii) evaluated the performance of the scheme on different sets of C. perfringens genomes from different hosts and geographic regions as well as from different outbreak situations, and, finally, (iii) made this scheme publicly available supported by an allele nomenclature database for global and standard genomic typing.
Keywords: Clostridium perfringens; SNP; cgMLST; genome typing.
Conflict of interest statement
D.H. is one of the owners of the proprietary software SeqSphere+ that was used in different analysis steps in this article. All other authors declare that there are no conflicts of interest.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

