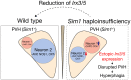
Ectopic expression of Irx3 and Irx5 in the paraventricular nucleus of the hypothalamus contributes to defects in Sim1 haploinsufficiency
- PMID: 34705510
- PMCID: PMC8550250
- DOI: 10.1126/sciadv.abh4503
Ectopic expression of Irx3 and Irx5 in the paraventricular nucleus of the hypothalamus contributes to defects in Sim1 haploinsufficiency
Abstract
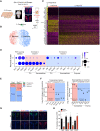
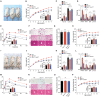
The paraventricular nucleus of the hypothalamus (PVH) contains a heterogeneous cluster of Sim1-expressing neurons critical for feeding regulation. Sim1 haploinsufficiency results in hyperphagic obesity with disruption of PVH neurons, yet the molecular profiles of PVH neurons and the mechanism underlying the defects of Sim1 haploinsufficiency are not well understood. By single-cell RNA sequencing, we identified two major populations of Sim1+ PVH neurons, which are differentially affected by Sim1 haploinsufficiency. The Iroquois homeobox genes Irx3 and Irx5 have been implicated in the hypothalamic control of energy homeostasis. We found that Irx3 and Irx5 are ectopically expressed in the Sim1+ PVH cells of Sim1+/− mice. By reducing their dosage and PVH-specific deletion of Irx3, we demonstrate that misexpression of Irx3 and Irx5 contributes to the defects of Sim1+/− mice. Our results illustrate abnormal hypothalamic activities of Irx3 and Irx5 as a central mechanism disrupting PVH development and feeding regulation in Sim1 haploinsufficiency.
Figures




Similar articles
-
Control of food intake and energy expenditure by Nos1 neurons of the paraventricular hypothalamus.J Neurosci. 2014 Nov 12;34(46):15306-18. doi: 10.1523/JNEUROSCI.0226-14.2014. J Neurosci. 2014. PMID: 25392498 Free PMC article.
-
Irx3 and Irx5 - Novel Regulatory Factors of Postnatal Hypothalamic Neurogenesis.Front Neurosci. 2021 Nov 2;15:763856. doi: 10.3389/fnins.2021.763856. eCollection 2021. Front Neurosci. 2021. PMID: 34795556 Free PMC article. Review.
-
Dnmt3a in Sim1 neurons is necessary for normal energy homeostasis.J Neurosci. 2014 Nov 12;34(46):15288-96. doi: 10.1523/JNEUROSCI.1316-14.2014. J Neurosci. 2014. PMID: 25392496 Free PMC article.
-
Sim1 haploinsufficiency impairs melanocortin-mediated anorexia and activation of paraventricular nucleus neurons.Mol Endocrinol. 2006 Oct;20(10):2483-92. doi: 10.1210/me.2005-0483. Epub 2006 May 25. Mol Endocrinol. 2006. PMID: 16728530
-
The Paraventricular Nucleus of the Hypothalamus: Development, Function, and Human Diseases.Endocrinology. 2018 Sep 1;159(9):3458-3472. doi: 10.1210/en.2018-00453. Endocrinology. 2018. PMID: 30052854 Review.
Cited by
-
The Iroquois (Iro/Irx) homeobox genes are conserved Hox targets involved in motor neuron development.bioRxiv [Preprint]. 2024 May 30:2024.05.30.596714. doi: 10.1101/2024.05.30.596714. bioRxiv. 2024. Update in: iScience. 2025 Mar 12;28(4):112210. doi: 10.1016/j.isci.2025.112210. PMID: 38853975 Free PMC article. Updated. Preprint.
-
Genetics, pharmacotherapy, and dietary interventions in childhood obesity.J Pharm Pharm Sci. 2024 May 28;27:12861. doi: 10.3389/jpps.2024.12861. eCollection 2024. J Pharm Pharm Sci. 2024. PMID: 38863827 Free PMC article. Review.
-
Unlocking biological mechanisms with integrative functional genomics approaches.Mol Cells. 2024 Aug;47(8):100092. doi: 10.1016/j.mocell.2024.100092. Epub 2024 Jul 15. Mol Cells. 2024. PMID: 39019219 Free PMC article. Review.
-
The paraventricular nucleus of the hypothalamus: a key node in the control of behavioural states.J Physiol. 2025 Apr;603(8):2231-2243. doi: 10.1113/JP288366. Epub 2025 Mar 22. J Physiol. 2025. PMID: 40119815 Free PMC article. Review.
-
A Spacetime Odyssey of Neural Progenitors to Generate Neuronal Diversity.Neurosci Bull. 2023 Apr;39(4):645-658. doi: 10.1007/s12264-022-00956-0. Epub 2022 Oct 10. Neurosci Bull. 2023. PMID: 36214963 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

