Genomic Analysis of a Hospital-Associated Outbreak of Mycobacterium abscessus: Implications on Transmission
- PMID: 34705540
- PMCID: PMC8769749
- DOI: 10.1128/JCM.01547-21
Genomic Analysis of a Hospital-Associated Outbreak of Mycobacterium abscessus: Implications on Transmission
Abstract
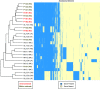
Whole-genome sequencing (WGS) has recently been used to investigate acquisition of Mycobacterium abscessus. Investigators have reached conflicting conclusions about the meaning of genetic distances for interpretation of person-to-person transmission. Existing genomic studies were limited by a lack of WGS from environmental M. abscessus isolates. In this study, we retrospectively analyzed the core and accessory genomes of 26 M. abscessus subsp. abscessus isolates collected over 7 years. Clinical isolates (n = 22) were obtained from a large hospital-associated outbreak of M. abscessus subsp. abscessus, the outbreak hospital before or after the outbreak, a neighboring hospital, and two outside laboratories. Environmental M. abscessus subsp. abscessus isolates (n = 4) were obtained from outbreak hospital water outlets. Phylogenomic analysis of study isolates revealed three clades with pairwise genetic distances ranging from 0 to 135 single-nucleotide polymorphisms (SNPs). Compared to a reference environmental outbreak isolate, all seven clinical outbreak isolates and the remaining three environmental isolates had highly similar core and accessory genomes, differing by up to 7 SNPs and a median of 1.6% accessory genes, respectively. Although genomic comparisons of 15 nonoutbreak clinical isolates revealed greater heterogeneity, five (33%) isolates had fewer than 20 SNPs compared to the reference environmental isolate, including two unrelated outside laboratory isolates with less than 4% accessory genome variation. Detailed genomic comparisons confirmed environmental acquisition of outbreak isolates of M. abscessus subsp. abscessus. SNP distances alone, however, did not clearly differentiate the mechanism of acquisition of outbreak versus nonoutbreak isolates. We conclude that successful investigation of M. abscessus subsp. abscessus clusters requires molecular and epidemiologic components, ideally complemented by environmental sampling.
Keywords: Mycobacterium abscessus; hospital outbreak; infection prevention; nontuberculous mycobacteria; whole-genome sequencing.
Figures


References
-
- Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Jr, Winthrop K, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. 10.1164/rccm.200604-571ST. - DOI - PubMed
-
- Pasipanodya JG, Ogbonna D, Ferro BE, Magombedze G, Srivastava S, Deshpande D, Gumbo T. 2017. Systematic review and meta-analyses of the effect of chemotherapy on pulmonary Mycobacterium abscessus outcomes and disease recurrence. Antimicrob Agents Chemother 61:e01206-17. 10.1128/AAC.01206-17. - DOI - PMC - PubMed
-
- Floto RA, Olivier KN, Saiman L, Daley CL, Herrmann JL, Nick JA, Noone PG, Bilton D, Corris P, Gibson RL, Hempstead SE, Koetz K, Sabadosa KA, Sermet-Gaudelus I, Smyth AR, van Ingen J, Wallace RJ, Winthrop KL, Marshall BC, Haworth CS, US Cystic Fibrosis Foundation and European Cystic Fibrosis Society. 2016. US Cystic Fibrosis Foundation and European Cystic Fibrosis Society consensus recommendations for the management of non-tuberculous mycobacteria in individuals with cystic fibrosis. Thorax 71(Suppl 1):i1–i22. 10.1136/thoraxjnl-2015-207360. - DOI - PMC - PubMed
-
- Alcaide F, Pena MJ, Perez-Risco D, Camprubi D, Gonzalez-Luquero L, Grijota-Camino MD, Dorca J, Santin M. 2017. Increasing isolation of rapidly growing mycobacteria in a low-incidence setting of environmental mycobacteria, 1994-2015. Eur J Clin Microbiol Infect Dis 36:1425–1432. 10.1007/s10096-017-2949-0. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

