A Novel Actinobacterial Cutinase Containing a Noncatalytic Polymer-Binding Domain
- PMID: 34705546
- PMCID: PMC8752134
- DOI: 10.1128/AEM.01522-21
A Novel Actinobacterial Cutinase Containing a Noncatalytic Polymer-Binding Domain
Abstract
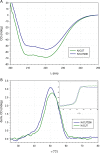
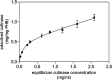
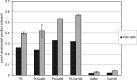
The single putative cutinase-encoding gene from the genome of Kineococcus radiotolerans SRS30216 was cloned and expressed in Escherichia coli as a secreted fusion protein, designated YebF-KrCUT, where YebF is the extracellular carrier protein. The 294-amino-acid sequence of KrCUT is unique among currently characterized cutinases by having a C-terminal extension that consists of a short (Pro-Thr)-rich linker and a 55-amino-acid region resembling the substrate binding domain of poly(hydroxybutyrate) (PHB) depolymerases. Phylogenetically, KrCUT takes a unique position among known cutinases and cutinase-like proteins of bacterial and fungal origins. A modeled structure of KrCUT, although displaying a typical α/β hydrolase fold, shows some unique loops close to the catalytic site. The 39-kDa YebF-KrCUT fusion protein and a truncated variant thereof were purified to electrophoretic homogeneity and functionally characterized. The melting temperatures (Tm) of KrCUT and its variant KrCUT206 devoid of the putative PHB-binding domain were established to be very similar, at 50 to 51°C. Cutinase activity was confirmed by the appearance of characteristic cutin components, C16 and C18 hydroxyl fatty acids, in the mass chromatograms following incubation of KrCUT with apple cutin as the substrate. KrCUT also efficiently degraded synthetic polyesters such as polycaprolactone and poly(1,3-propylene adipate). Although incapable of PHB depolymerization, KrCUT could efficiently bind PHB, confirming the predicted characteristic of the C-terminal region. KrCUT also potentiated the activity of pectate lyase in the degradation of pectin from hemp fibers. This synergistic effect is relevant to the enzyme retting process of natural fibers. IMPORTANCE To date, only a limited number of cutinases have been isolated and characterized from nature, the majority being sourced from phytopathogenic fungi and thermophilic bacteria. The significance of our research relates to the identification and characterization of a unique member of the microbial cutinases, named KrCUT, that was derived from the genome of the Gram-positive Kineococcus radiotolerans SRS30216, a highly radiation-resistant actinobacterium. Given the wide-ranging importance of cutinases in applications such as the degradation of natural and synthetic polymers, in the textile industry, in laundry detergents, and in biocatalysis (e.g., transesterification reactions), our results could foster new research leading to broader biotechnological impacts. This study also demonstrated that genome mining or prospecting is a viable means to discover novel biocatalysts as environmentally friendly and biotechnological tools.
Keywords: Kineococcus; biopolymer degradation; biotechnology; cutinase; enzyme technology; microbial world; α/β fold hydrolase.
Figures





References
-
- Riederer M, Mueller C. 2006. Annual plant reviews, vol 23. Biology of the plant cuticle. Blackwell Publishing Ltd, Oxford, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

