ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress
- PMID: 34706264
- PMCID: PMC8502680
- DOI: 10.1016/j.devcel.2021.10.006
ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress
Abstract
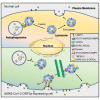
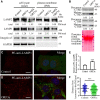
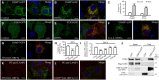
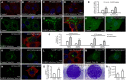
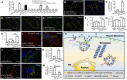
Viral entry and egress are important determinants of virus infectivity and pathogenicity. β-coronaviruses, including the COVID-19 virus SARS-CoV-2 and mouse hepatitis virus (MHV), exploit the lysosomal exocytosis pathway for egress. Here, we show that SARS-CoV-2 ORF3a, but not SARS-CoV ORF3a, promotes lysosomal exocytosis. SARS-CoV-2 ORF3a facilitates lysosomal targeting of the BORC-ARL8b complex, which mediates trafficking of lysosomes to the vicinity of the plasma membrane, and exocytosis-related SNARE proteins. The Ca2+ channel TRPML3 is required for SARS-CoV-2 ORF3a-mediated lysosomal exocytosis. Expression of SARS-CoV-2 ORF3a greatly elevates extracellular viral release in cells infected with the coronavirus MHV-A59, which itself lacks ORF3a. In SARS-CoV-2 ORF3a, Ser171 and Trp193 are critical for promoting lysosomal exocytosis and blocking autophagy. When these residues are introduced into SARS-CoV ORF3a, it acquires the ability to promote lysosomal exocytosis and inhibit autophagy. Our results reveal a mechanism by which SARS-CoV-2 interacts with host factors to promote its extracellular egress.
Keywords: COVID-19; ORF3a; SARS-CoV; SARS-CoV-2; lysosomal exocytosis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Braulke T., Bonifacino J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta. 2009;1793:605–614. - PubMed
-
- Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., et al. Role of severe acute respiratory syndrome coronavirus Viroporins E, 3a, and 8a in Replication and pathogenesis. mBio. 2018;9 e02325–17. - PMC - PubMed
-
- Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R., et al. The open reading frame 3a protein of severe acute respiratory syndrome- associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84:1097–1109. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

