A small number of early introductions seeded widespread transmission of SARS-CoV-2 in Québec, Canada
- PMID: 34706766
- PMCID: PMC8550813
- DOI: 10.1186/s13073-021-00986-9
A small number of early introductions seeded widespread transmission of SARS-CoV-2 in Québec, Canada
Abstract
Background: Québec was the Canadian province most impacted by COVID-19, with 401,462 cases as of September 24th, 2021, and 11,347 deaths due mostly to a very severe first pandemic wave. In April 2020, we assembled the Coronavirus Sequencing in Québec (CoVSeQ) consortium to sequence SARS-CoV-2 genomes in Québec to track viral introduction events and transmission within the province.
Methods: Using genomic epidemiology, we investigated the arrival of SARS-CoV-2 to Québec. We report 2921 high-quality SARS-CoV-2 genomes in the context of > 12,000 publicly available genomes sampled globally over the first pandemic wave (up to June 1st, 2020). By combining phylogenetic and phylodynamic analyses with epidemiological data, we quantify the number of introduction events into Québec, identify their origins, and characterize the spatiotemporal spread of the virus.
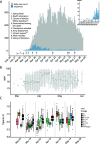
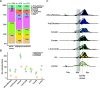
Results: Conservatively, we estimated approximately 600 independent introduction events, the majority of which happened from spring break until 2 weeks after the Canadian border closed for non-essential travel. Subsequent mass repatriations did not generate large transmission lineages (> 50 sequenced cases), likely due to mandatory quarantine measures in place at the time. Consistent with common spring break and "snowbird" destinations, most of the introductions were inferred to have originated from Europe via the Americas. Once introduced into Québec, viral lineage sizes were overdispersed, with a few lineages giving rise to most infections. Consistent with founder effects, the earliest lineages to arrive tended to spread most successfully. Fewer than 100 viral introductions arrived during spring break, of which 7-12 led to the largest transmission lineages of the first wave (accounting for 52-75% of all sequenced infections). These successful transmission lineages dispersed widely across the province. Transmission lineage size was greatly reduced after March 11th, when a quarantine order for returning travellers was enacted. While this suggests the effectiveness of early public health measures, the biggest transmission lineages had already been ignited prior to this order.
Conclusions: Combined, our results reinforce how, in the absence of tight travel restrictions or quarantine measures, fewer than 100 viral introductions in a week can ensure the establishment of extended transmission chains.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Komissarov AB, Safina KR, Garushyants SK, Fadeev AV, Sergeeva MV, Ivanova AA, Danilenko DM, Lioznov D, Shneider OV, Shvyrev N, Spirin V, Glyzin D, Shchur V, Bazykin GA. Genomic epidemiology of the early stages of the SARS-CoV-2 outbreak in Russia. Nat Commun. 2021;12(1):649. doi: 10.1038/s41467-020-20880-z. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

