MicroRNA-206 promotes the recruitment of CD8+ T cells by driving M1 polarisation of Kupffer cells
- PMID: 34706869
- PMCID: PMC9279850
- DOI: 10.1136/gutjnl-2021-324170
MicroRNA-206 promotes the recruitment of CD8+ T cells by driving M1 polarisation of Kupffer cells
Abstract
Objective: Kupffer cells (KCs) protect against hepatocellular carcinoma (HCC) by communicating with other immune cells. However, the underlying mechanism(s) of this process is incompletely understood.
Design: FVB/NJ mice were hydrodynamically injected with AKT/Ras and Sleeping Beauty transposon to induce HCC. Mini-circle and Sleeping Beauty were used to overexpress microRNA-206 in KCs of mice. Flow cytometry and immunostaining were used to evaluate the change in the immune system.
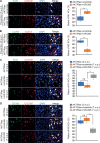
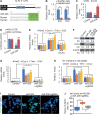
Results: Hydrodynamic injection of AKT/Ras into mice drove M2 polarisation of KCs and depletion of cytotoxic T cells (CTLs) and promoted HCC development. M1-to-M2 transition of KCs impaired microRNA-206 biogenesis. By targeting Klf4 (kruppel like factor 4) and, thereby, enhancing the production of M1 markers including C-C motif chemokine ligand 2 (CCL2), microRNA-206 promoted M1 polarisation of macrophages. Indeed, microRNA-206-mediated increase of CCL2 facilitated hepatic recruitment of CTLs via CCR2. Disrupting each component of the KLF4/CCL2/CCR2 axis impaired the ability of microRNA-206 to drive M1 polarisation of macrophages and recruit CTLs. In AKT/Ras mice, KC-specific expression of microRNA-206 drove M1 polarisation of KCs and hepatic recruitment of CTLs and fully prevented HCC, while 100% of control mice died from HCC. Disrupting the interaction between microRNA-206 and Klf4 in KCs and depletion of CD8+ T cells impaired the ability of miR-206 to prevent HCC.
Conclusions: M2 polarisation of KCs is a major contributor of HCC in AKT/Ras mice. MicroRNA-206, by driving M1 polarisation of KCs, promoted the recruitment of CD8+ T cells and prevented HCC, suggesting its potential use as an immunotherapeutic approach.
Keywords: Kupffer cell; hepatocellular carcinoma; immunotherapy.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
