Cryo-EM structure of the needle filament tip complex of the Salmonella type III secretion injectisome
- PMID: 34706941
- PMCID: PMC8612237
- DOI: 10.1073/pnas.2114552118
Cryo-EM structure of the needle filament tip complex of the Salmonella type III secretion injectisome
Abstract
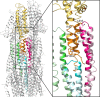
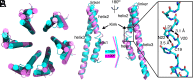
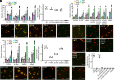
Type III secretion systems are multiprotein molecular machines required for the virulence of several important bacterial pathogens. The central element of these machines is the injectisome, a ∼5-Md multiprotein structure that mediates the delivery of bacterially encoded proteins into eukaryotic target cells. The injectisome is composed of a cytoplasmic sorting platform, and a membrane-embedded needle complex, which is made up of a multiring base and a needle-like filament that extends several nanometers from the bacterial surface. The needle filament is capped at its distal end by another substructure known as the tip complex, which is crucial for the translocation of effector proteins through the eukaryotic cell plasma membrane. Here we report the cryo-EM structure of the Salmonella Typhimurium needle tip complex docked onto the needle filament tip. Combined with a detailed analysis of structurally guided mutants, this study provides major insight into the assembly and function of this essential component of the type III secretion protein injection machine.
Keywords: bacterial pathogenesis; cryoelectron microscopy; organelle assembly; protein secretion; structural biology.
Conflict of interest statement
The authors declare no competing interest.
Figures
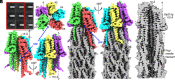


References
-
- Deng W., et al., Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

