The orbitofrontal cortex maps future navigational goals
- PMID: 34707289
- PMCID: PMC8599015
- DOI: 10.1038/s41586-021-04042-9
The orbitofrontal cortex maps future navigational goals
Abstract
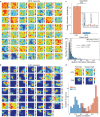
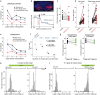
Accurate navigation to a desired goal requires consecutive estimates of spatial relationships between the current position and future destination throughout the journey. Although neurons in the hippocampal formation can represent the position of an animal as well as its nearby trajectories1-7, their role in determining the destination of the animal has been questioned8,9. It is, thus, unclear whether the brain can possess a precise estimate of target location during active environmental exploration. Here we describe neurons in the rat orbitofrontal cortex (OFC) that form spatial representations persistently pointing to the subsequent goal destination of an animal throughout navigation. This destination coding emerges before the onset of navigation, without direct sensory access to a distal goal, and even predicts the incorrect destination of an animal at the beginning of an error trial. Goal representations in the OFC are maintained by destination-specific neural ensemble dynamics, and their brief perturbation at the onset of a journey led to a navigational error. These findings suggest that the OFC is part of the internal goal map of the brain, enabling animals to navigate precisely to a chosen destination that is beyond the range of sensory perception.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

