Potential Common Genetic Risks of Sporadic Parkinson's Disease and Amyotrophic Lateral Sclerosis in the Han Population of Mainland China
- PMID: 34707478
- PMCID: PMC8542930
- DOI: 10.3389/fnins.2021.753870
Potential Common Genetic Risks of Sporadic Parkinson's Disease and Amyotrophic Lateral Sclerosis in the Han Population of Mainland China
Abstract
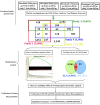
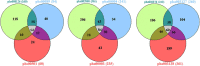
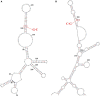
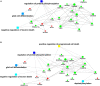
Sporadic Parkinson's disease (sPD) and sporadic amyotrophic lateral sclerosis (sALS) are neurodegenerative diseases characterized by progressive and selective neuron death, with some genetic similarities. In order to investigate the genetic risk factors common to both sPD and sALS, we carried out a screen of risk alleles for sALS and related loci in 530 sPD patients and 530 controls from the Han population of Mainland China (HPMC). We selected 27 single-nucleotide polymorphisms in 10 candidate genes associated with sALS, and we performed allelotyping and genotyping to determine their frequencies in the study population as well as bioinformatics analysis to assess their functional significance in these diseases. The minor alleles of rs17115303 in DAB adaptor protein 1 (DAB1) gene and rs6030462 in protein tyrosine phosphatase receptor type T (PTPRT) gene were correlated with increased risk of both sPD and sALS. Polymorphisms of rs17115303 and rs6030462 were associated with alterations in transcription factor binding sites, secondary structures, long non-coding RNA interactions, and nervous system regulatory networks; these changes involved biological processes associated with neural cell development, differentiation, neurogenesis, migration, axonogenesis, cell adhesion, and metabolism of phosphate-containing compounds. Thus, variants of DAB1 gene (rs17115303) and PTPRT gene (rs6030462) are risk factors common to sPD and sALS in the HPMC. These findings provide insight into the molecular pathogenesis of both diseases and can serve as a basis for the development of targeted therapies.
Keywords: amyotrophic lateral sclerosis; common genetic risk; pathogenesis; polymorphism; sporadic Parkinson’s disease.
Copyright © 2021 Lu, Chen, Wei, Zhu and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
The rs2619566, rs10260404, and rs79609816 Polymorphisms Are Associated With Sporadic Amyotrophic Lateral Sclerosis in Individuals of Han Ancestry From Mainland China.Front Genet. 2021 Aug 6;12:679204. doi: 10.3389/fgene.2021.679204. eCollection 2021. Front Genet. 2021. PMID: 34421992 Free PMC article.
-
Association analysis of polymorphisms in VMAT2 and TMEM106B genes for Parkinson's disease, amyotrophic lateral sclerosis and multiple system atrophy.J Neurol Sci. 2017 Jun 15;377:65-71. doi: 10.1016/j.jns.2017.03.028. Epub 2017 Mar 21. J Neurol Sci. 2017. PMID: 28477711
-
Polymorphisms in protein disulfide isomerase are associated with sporadic amyotrophic lateral sclerosis in the Chinese Han population.Int J Neurosci. 2016;126(7):607-11. doi: 10.3109/00207454.2015.1050098. Epub 2015 Aug 18. Int J Neurosci. 2016. PMID: 26000911
-
Sporadic Parkinson's Disease Potential Risk Loci Identified in Han Ancestry of Chinese Mainland.Front Aging Neurosci. 2021 Jan 12;12:603793. doi: 10.3389/fnagi.2020.603793. eCollection 2020. Front Aging Neurosci. 2021. PMID: 33510632 Free PMC article.
-
Sporadic amyotrophic lateral sclerosis: a hypothesis of persistent (non-lytic) enteroviral infection.Amyotroph Lateral Scler Other Motor Neuron Disord. 2005 Jun;6(2):77-87. doi: 10.1080/14660820510027026. Amyotroph Lateral Scler Other Motor Neuron Disord. 2005. PMID: 16036430 Review.
Cited by
-
Hereditable variants of classical protein tyrosine phosphatase genes: Will they prove innocent or guilty?Front Cell Dev Biol. 2023 Jan 23;10:1051311. doi: 10.3389/fcell.2022.1051311. eCollection 2022. Front Cell Dev Biol. 2023. PMID: 36755664 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

