A brief history and popularity of methods and tools used to estimate micro-evolutionary forces
- PMID: 34707813
- PMCID: PMC8525119
- DOI: 10.1002/ece3.8076
A brief history and popularity of methods and tools used to estimate micro-evolutionary forces
Abstract
Population genetics is a field of research that predates the current generations of sequencing technology. Those approaches, that were established before massively parallel sequencing methods, have been adapted to these new marker systems (in some cases involving the development of new methods) that allow genome-wide estimates of the four major micro-evolutionary forces-mutation, gene flow, genetic drift, and selection. Nevertheless, classic population genetic markers are still commonly used and a plethora of analysis methods and programs is available for these and high-throughput sequencing (HTS) data. These methods employ various and diverse theoretical and statistical frameworks, to varying degrees of success, to estimate similar evolutionary parameters making it difficult to get a concise overview across the available approaches. Presently, reviews on this topic generally focus on a particular class of methods to estimate one or two evolutionary parameters. Here, we provide a brief history of methods and a comprehensive list of available programs for estimating micro-evolutionary forces. We furthermore analyzed their usage within the research community based on popularity (citation bias) and discuss the implications of this bias for the software community. We found that a few programs received the majority of citations, with program success being independent of both the parameters estimated and the computing platform. The only deviation from a model of exponential growth in the number of citations was found for the presence of a graphical user interface (GUI). Interestingly, no relationship was found for the impact factor of the journals, when the tools were published, suggesting accessibility might be more important than visibility.
Keywords: drift; migration; mutation; population genetics; selection; software; user bias.
© 2021 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
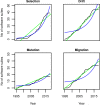

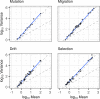

References
-
- Àlvarez, D. , Lourenço, A. , Oro, D. , & Veló‐Anton, G. (2015). Assessment of census (N) and effective population size (Ne) reveals consistency of Ne single‐sample estimators and a high Ne/N ratio in an urban and isolated population of fire salamander. Conservation Genetic Resources, 7, 705–712.
Publication types
Associated data
LinkOut - more resources
Full Text Sources

