Population pharmacokinetic model selection assisted by machine learning
- PMID: 34708337
- PMCID: PMC8940812
- DOI: 10.1007/s10928-021-09793-6
Population pharmacokinetic model selection assisted by machine learning
Abstract
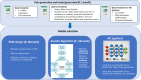
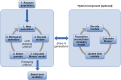
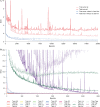
A fit-for-purpose structural and statistical model is the first major requirement in population pharmacometric model development. In this manuscript we discuss how this complex and computationally intensive task could benefit from supervised machine learning algorithms. We compared the classical pharmacometric approach with two machine learning methods, genetic algorithm and neural networks, in different scenarios based on simulated pharmacokinetic data. Genetic algorithm performance was assessed using a fitness function based on log-likelihood, whilst neural networks were trained using mean square error or binary cross-entropy loss. Machine learning provided a selection based only on statistical rules and achieved accurate selection. The minimization process of genetic algorithm was successful at allowing the algorithm to select plausible models. Neural network classification tasks achieved the most accurate results. Neural network regression tasks were less precise than neural network classification and genetic algorithm methods. The computational gain obtained by using machine learning was substantial, especially in the case of neural networks. We demonstrated that machine learning methods can greatly increase the efficiency of pharmacokinetic population model selection in case of large datasets or complex models requiring long run-times. Our results suggest that machine learning approaches can achieve a first fast selection of models which can be followed by more conventional pharmacometric approaches.
Keywords: Deep learning; Genetic algorithm; Model-informed drug discovery and development; Neural network; Pharmacometrics; Population PK/PD.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources

