Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection
- PMID: 34709128
- PMCID: PMC8635679
- DOI: 10.1080/22221751.2021.1999779
Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection
Abstract
The Georgia-07-like genotype II African swine fever virus (ASFV) with high virulence has been prevalent in China since 2018. Here, we report that genotype I ASFVs have now also emerged in China. Two non-haemadsorbing genotype I ASFVs, HeN/ZZ-P1/21 and SD/DY-I/21, were isolated from pig farms in Henan and Shandong province, respectively. Phylogenetic analysis of the whole genome sequences suggested that both isolates share high similarity with NH/P68 and OURT88/3, two genotype I ASFVs isolated in Portugal in the last century. Animal challenge testing revealed that SD/DY-I/21 shows low virulence and efficient transmissibility in pigs, and causes mild onset of infection and chronic disease. SD/DY-I/21 was found to cause necrotic skin lesions and joint swelling. The emergence of genotype I ASFVs will present more problems and challenges for the control and prevention of African swine fever in China.
Keywords: African swine fever virus; China; genotype I; pig; virulence.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




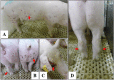

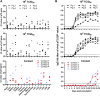
References
-
- Wardley RC, et al. . African swine fever virus. Brief review. Arch Virol. 1983;76(2):73–90. - PubMed
-
- Dixon LK, Sun H, Roberts H.. African swine fever. Antiviral Res. 2019;165:34–41. - PubMed
-
- Muangkram Y, Sukmak M, Wajjwalku W.. Phylogeographic analysis of African swine fever virus based on the p72 gene sequence. Genet Mol Res. 2015;14(2):4566–4574. - PubMed
-
- Gao L, et al. . Epidemic situation and control measures of African Swine Fever Outbreaks in China 2018-2020. Transbound Emerg Dis. 2021;68(5):2676–2686. - PubMed
-
- Penrith ML, et al. . African swine fever virus eradication in Africa. Virus Res. 2013;173(1):228–246. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
