Molecular characterization, receptor binding property, and replication in chickens and mice of H9N2 avian influenza viruses isolated from chickens, peafowls, and wild birds in eastern China
- PMID: 34709136
- PMCID: PMC8592596
- DOI: 10.1080/22221751.2021.1999778
Molecular characterization, receptor binding property, and replication in chickens and mice of H9N2 avian influenza viruses isolated from chickens, peafowls, and wild birds in eastern China
Abstract
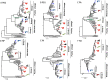
H9N2 avian influenza viruses are widely prevalent in birds and pose an increasing threat to humans because of their enhanced virulence and transmissibility in mammals. Active surveillance on the prevalence and evolution of H9N2 viruses in different avian hosts will help develop eradication measures. We isolated 16 H9N2 viruses from chickens, green peafowls, and wild birds in eastern China from 2017 to 2019 and characterized their comparative genetic evolution, receptor-binding specificity, antigenic diversity, replication, and transmission in chickens and mice. The phylogenetic analysis indicated that the green peafowl viruses and swan reassortant shared the same ancestor with the poultry H9N2 viruses prevalent in eastern China, while the seven wild bird viruses belonged to wild bird lineage. The chicken, peafowl, and swan H9N2 viruses that belonged to the poultry lineage preferentially recognized α-2, 6-linked sialic acids (human-like receptor), but the wild bird lineage viruses can bind both α-2, 3 (avian-like receptor) and human-like receptor similarly. Interestingly, the H9N2 viruses of poultry lineage replicated well and transmitted efficiently, but the viruses of wild bird lineage replicated and transmitted with low efficiency. Importantly, the H9N2 viruses of poultry lineage replicated in higher titer in mammal cells and mice than the viruses of wild birds lineage. Altogether, our study indicates that co-circulation of the H9N2 viruses in poultry, wild birds, and ornamental birds increased their cross-transmission risk in different birds because of their widespread dissemination.
Keywords: Avian influenza virus; H9N2; chicken; peafowl; wild birds.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Shi JZ, Deng GH, Liu PH, et al. Isolation and characterization of H7N9 viruses from live poultry markets-implication of the source of current H7N9 infection in humans. Chinese Sci Bull. 2013 Jun;58(16):1857–1863. doi: 10.1007/s11434-013-5873-4. PubMed PMID: WOS:000319850700003; English. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
