Disrupted PGR-B and ESR1 signaling underlies defective decidualization linked to severe preeclampsia
- PMID: 34709177
- PMCID: PMC8553341
- DOI: 10.7554/eLife.70753
Disrupted PGR-B and ESR1 signaling underlies defective decidualization linked to severe preeclampsia
Abstract
Background: Decidualization of the uterine mucosa drives the maternal adaptation to invasion by the placenta. Appropriate depth of placental invasion is needed to support a healthy pregnancy; shallow invasion is associated with the development of severe preeclampsia (sPE). Maternal contribution to sPE through failed decidualization is an important determinant of placental phenotype. However, the molecular mechanism underlying the in vivo defect linking decidualization to sPE is unknown.
Methods: Global RNA sequencing was applied to obtain the transcriptomic profile of endometrial biopsies collected from nonpregnant women who suffer sPE in a previous pregnancy and women who did not develop this condition. Samples were randomized in two cohorts, the training and the test set, to identify the fingerprinting encoding defective decidualization in sPE and its subsequent validation. Gene Ontology enrichment and an interaction network were performed to deepen in pathways impaired by genetic dysregulation in sPE. Finally, the main modulators of decidualization, estrogen receptor 1 (ESR1) and progesterone receptor B (PGR-B), were assessed at the level of gene expression and protein abundance.
Results: Here, we discover the footprint encoding this decidualization defect comprising 120 genes-using global gene expression profiling in decidua from women who developed sPE in a previous pregnancy. This signature allowed us to effectively segregate samples into sPE and control groups. ESR1 and PGR were highly interconnected with the dynamic network of the defective decidualization fingerprint. ESR1 and PGR-B gene expression and protein abundance were remarkably disrupted in sPE.
Conclusions: Thus, the transcriptomic signature of impaired decidualization implicates dysregulated hormonal signaling in the decidual endometria in women who developed sPE. These findings reveal a potential footprint that could be leveraged for a preconception or early prenatal screening of sPE risk, thus improving prevention and early treatments.
Funding: This work has been supported by the grant PI19/01659 (MCIU/AEI/FEDER, UE) from the Spanish Carlos III Institute awarded to TGG. NCM was supported by the PhD program FDGENT/2019/008 from the Spanish Generalitat Valenciana. IMB was supported by the PhD program PRE2019-090770 and funding was provided by the grant RTI2018-094946-B-100 (MCIU/AEI/FEDER, UE) from the Spanish Ministry of Science and Innovation with CS as principal investigator. This research was funded partially by Igenomix S.L.
Keywords: Preeclampsia; chromosomes; decidua; defective decidualization; gene expression; hormonal receptor; human; human endometrium; medicine; transcriptomic fingerprint.
© 2021, Garrido-Gomez et al.
Conflict of interest statement
TG, NC, MC, TC, IM, AA, JJ, RM, RC, AP, CS No competing interests declared
Figures
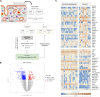
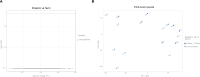


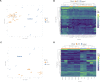

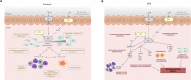

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

