p21 produces a bioactive secretome that places stressed cells under immunosurveillance
- PMID: 34709885
- PMCID: PMC8985214
- DOI: 10.1126/science.abb3420
p21 produces a bioactive secretome that places stressed cells under immunosurveillance
Abstract
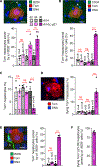
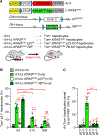
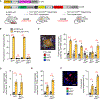
Immune cells identify and destroy damaged cells to prevent them from causing cancer or other pathologies by mechanisms that remain poorly understood. Here, we report that the cell-cycle inhibitor p21 places cells under immunosurveillance to establish a biological timer mechanism that controls cell fate. p21 activates retinoblastoma protein (Rb)–dependent transcription at select gene promoters to generate a complex bioactive secretome, termed p21-activated secretory phenotype (PASP). The PASP includes the chemokine CXCL14, which promptly attracts macrophages. These macrophages disengage if cells normalize p21 within 4 days, but if p21 induction persists, they polarize toward an M1 phenotype and lymphocytes mount a cytotoxic T cell response to eliminate target cells, including preneoplastic cells. Thus, p21 concurrently induces proliferative arrest and immunosurveillance of cells under duress.
Figures




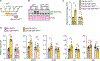
Comment in
-
Clearing stressed cells.Science. 2021 Oct 29;374(6567):534-535. doi: 10.1126/science.abm3229. Epub 2021 Oct 28. Science. 2021. PMID: 34709889
References
-
- Galluzzi L, Yamazaki T, Kroemer G, Linking cellular stress responses to systemic homeostasis. Nat Rev Mol Cell Biol 19, 731–745 (2018). - PubMed
-
- Koren E, Fuchs Y, Modes of Regulated Cell Death in Cancer. Cancer discovery 11, 245–265 (2021). - PubMed
-
- Kang TW et al., Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

