Alterations of lung microbial communities in obese allergic asthma and metabolic potential
- PMID: 34710121
- PMCID: PMC8553092
- DOI: 10.1371/journal.pone.0256848
Alterations of lung microbial communities in obese allergic asthma and metabolic potential
Abstract
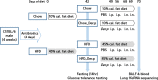
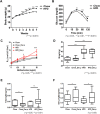
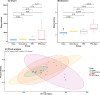
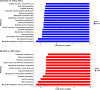
In recent years, there has been a rapid increase in microbiome studies to explore microbial alterations causing disease status and unveil disease pathogenesis derived from microbiome environmental modifications. Convincing evidence of lung microbial changes involving asthma has been collected; however, whether lung microbial changes under obesity leads to severe asthma in a state of allergen exposure has not been studied sufficiently. Here, we measured bacterial alterations in the lung of an allergen mouse model induced by a high fat diet (HFD) by using 16S rRNA gene sequencing. A total of 33 pathogen‑free 3‑week‑old male C57BL/6 mice were used, and they divided randomly into two groups. The Chow diet (n = 16) and high fat diet (n = 17) was administrated for 70 days. Mice were sensitized with PBS or Dermatophagoides pteronyssinus extract (Der.p), and concentration levels of total IgE and Der.p-IgE in the blood were measured to quantify immune responses. Although there were no meaningful differences in bacterial species richness in the HFD mouse group, momentous changes of bacterial diversity in the HFD mouse group were identified after the mouse group was exposed to allergens. At a genus level, the fluctuations of taxonomic relative abundances in several bacteria such as Ralstonia, Lactobacillus, Bradyrhizobium, Gaiella, PAC001932_g, Pseudolabrys, and Staphylococcus were conspicuously observed in the HFD mouse group exposed to allergens. Also, we predicted metabolic signatures occurring under microbial alterations in the Chow group versus the Chow group exposed to allergens, as well as in the HFD mouse group versus the HFD group exposed to allergens. We then compared their similarities and differences. Metabolic functions associated with macrophages such as propanoate metabolism, butanoate metabolism, and glycine-serine-threonine metabolism were identified in the HFD group versus the Chow group. These results provide new insights into the understanding of a microbiome community of obese allergic asthma, and shed light on the functional roles of lung microbiota inducing the pathogenesis of severe asthma.
Conflict of interest statement
No competing interests exist.
Figures