Genome-centric metagenomics reveals insights into the evolution and metabolism of a new free-living group in Rhizobiales
- PMID: 34711170
- PMCID: PMC8555084
- DOI: 10.1186/s12866-021-02354-4
Genome-centric metagenomics reveals insights into the evolution and metabolism of a new free-living group in Rhizobiales
Abstract
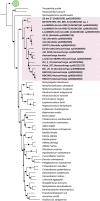
Background: The Rhizobiales (Proteobacteria) order is an abundant and diverse group of microorganisms, being extensively studied for its lifestyle based on the association with plants, animals, and humans. New studies have demonstrated that the last common ancestor (LCA) of Rhizobiales had a free-living lifestyle, but the phylogenetic and metabolism characterization of basal lineages remains unclear. Here, we used a high-resolution phylogenomic approach to test the monophyly of the Aestuariivirgaceae family, a new taxonomic group of Rhizobiales. Furthermore, a deep metabolic investigation provided an overview of the main functional traits that can be associated with its lifestyle. We hypothesized that the presence of pathways (e.g., Glycolysis/Gluconeogenesis) and the absence of pathogenic genes would be associated with a free-living lifestyle in Aestuariivirgaceae.
Results: Using high-resolution phylogenomics approaches, our results revealed a clear separation of Aestuariivirgaceae into a distinct clade of other Rhizobiales family, suggesting a basal split early group and corroborate the monophyly of this group. A deep functional annotation indicated a metabolic versatility, which includes putative genes related to sugar degradation and aerobic respiration. Furthermore, many of these traits could reflect a basal metabolism and adaptations of Rhizobiales, as such the presence of Glycolysis/Gluconeogenesis pathway and the absence of pathogenicity genes, suggesting a free-living lifestyle in the Aestuariivirgaceae members.
Conclusions: Aestuariivirgaceae (Rhizobiales) family is a monophyletic taxon of the Rhizobiales with a free-living lifestyle and a versatile metabolism that allows these microorganisms to survive in the most diverse microbiomes, demonstrating their adaptability to living in systems with different conditions, such as extremely cold environments to tropical rivers.
Keywords: Aestuariivirgaceae; Evolution; Integration of genomic public data; Rhizobiales; Uncultivated lineages.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

