miR548ai antagonism attenuates exosome-induced endothelial cell dysfunction
- PMID: 34711811
- PMCID: PMC8553949
- DOI: 10.1038/s41420-021-00720-9
miR548ai antagonism attenuates exosome-induced endothelial cell dysfunction
Erratum in
-
Correction: miR548ai antagonism attenuates exosome-induced endothelial cell dysfunction.Cell Death Discov. 2022 Sep 12;8(1):382. doi: 10.1038/s41420-022-01178-z. Cell Death Discov. 2022. PMID: 36096890 Free PMC article. No abstract available.
Abstract
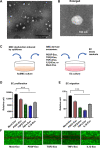
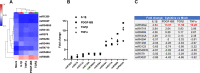
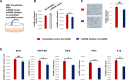
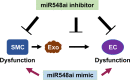
Endothelial cell (EC) and smooth muscle cell (SMC) are major cell types adjacent in the vascular wall. Recent progress indicates that their communication is crucial for vascular homeostasis and pathogenesis. In particular, dysfunctional (proliferative) SMCs through exosomes can induce EC dysfunction (impaired growth). The current study suggests that miR548ai, a rarely known microRNA, may provide a molecular target for protection against SMC/exosome-induced EC dysfunction. We performed microarray profiling of microRNAs of dysfunctional human primary aortic SMCs induced by different cytokines (PDGF-BB, TGFβ1, TNFα, IL1β). Among the microRNAs commonly upregulated by these cytokines, miR548ai showed the most robust changes, as also validated through quantitative PCR. This cytokine-induced miR548ai upregulation was recapitulated in the qPCR determination of SMC-derived exosomal microRNAs. Consistent with SMC-to-EC communication, the exosomes extracted from cytokine-stimulated SMCs impaired human EC proliferation and migration. Of particular interest, this SMC exosomal impingement on ECs was countered by transfection of miR548ai inhibitor microRNA into ECs. Furthermore, the miR548ai inhibitor transfected into SMCs attenuated SMC dysfunction/proliferation. Thus, these results identify miR548ai as a novel target; namely, miR548ai inhibitor mitigates EC dysfunction induced by exosomes derived from dysfunctional SMCs. This new knowledge may aid the future development of microRNA-based treatment of vascular disorders.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Grants and funding
- R01 HL129785/HL/NHLBI NIH HHS/United States
- R01 HL133665/HL/NHLBI NIH HHS/United States
- HL-133665/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- HL-129785/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R01 HL143469/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

