Scientific rationale for developing potent RBD-based vaccines targeting COVID-19
- PMID: 34711846
- PMCID: PMC8553742
- DOI: 10.1038/s41541-021-00393-6
Scientific rationale for developing potent RBD-based vaccines targeting COVID-19
Abstract
Vaccination of the global population against COVID-19 is a great scientific, logistical, and moral challenge. Despite the rapid development and authorization of several full-length Spike (S) protein vaccines, the global demand outweighs the current supply and there is a need for safe, potent, high-volume, affordable vaccines that can fill this gap, especially in low- and middle-income countries. Whether SARS-CoV-2 S-protein receptor-binding domain (RBD)-based vaccines could fill this gap has been debated, especially with regards to its suitability to protect against emerging viral variants of concern. Given a predominance for elicitation of neutralizing antibodies (nAbs) that target RBD following natural infection or vaccination, a key biomarker of protection, there is merit for selection of RBD as a sole vaccine immunogen. With its high-yielding production and manufacturing potential, RBD-based vaccines offer an abundance of temperature-stable doses at an affordable cost. In addition, as the RBD preferentially focuses the immune response to potent and recently recognized cross-protective determinants, this domain may be central to the development of future pan-sarbecovirus vaccines. In this study, we review the data supporting the non-inferiority of RBD as a vaccine immunogen compared to full-length S-protein vaccines with respect to humoral and cellular immune responses against both the prototype pandemic SARS-CoV-2 isolate and emerging variants of concern.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
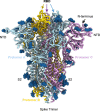
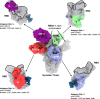
References
-
- WHO. Tracking SARS-CoV-2 Variants, www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (2021).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

