Transcriptomic analysis of loss of Gli1 in neural stem cells responding to demyelination in the mouse brain
- PMID: 34711861
- PMCID: PMC8553940
- DOI: 10.1038/s41597-021-01063-x
Transcriptomic analysis of loss of Gli1 in neural stem cells responding to demyelination in the mouse brain
Abstract
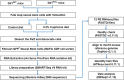
In the adult mammalian brain, Gli1 expressing neural stem cells reside in the subventricular zone and their progeny are recruited to sites of demyelination in the white matter where they generate new oligodendrocytes, the myelin forming cells. Remarkably, genetic loss or pharmacologic inhibition of Gli1 enhances the efficacy of remyelination by these neural stem cells. To understand the molecular mechanisms involved, we performed a transcriptomic analysis of this Gli1-pool of neural stem cells. We compared murine NSCs with either intact or deficient Gli1 expression from adult mice on a control diet or on a cuprizone diet which induces widespread demyelination. These data will be a valuable resource for identifying therapeutic targets for enhancing remyelination in demyelinating diseases like multiple sclerosis.
© 2021. The Author(s).
Conflict of interest statement
A patent on the method of targeting GLI1 as a strategy to promote remyelination has been awarded, with J. L. Salzer, J. Samanta and G. Fishell listed as co-inventors. JLS is a consultant for and has ownership interests in Glixogen Therapeutics.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

