The gastrointestinal nematodes of plains and Grevy's zebras: Phylogenetic relationships and host specificity
- PMID: 34712556
- PMCID: PMC8529100
- DOI: 10.1016/j.ijppaw.2021.10.007
The gastrointestinal nematodes of plains and Grevy's zebras: Phylogenetic relationships and host specificity
Abstract
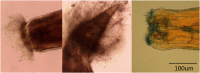
Equids are chronically infected with parasitic strongyle nematodes. There is a rich literature on horse strongyles, but they are difficult to identify morphologically and genetic studies on strongyles infecting other equid species are few, hampering studies of host specificity. We sequenced expelled worms from two sympatric zebra species in central Kenya to expand the strongyle phylogeny and used DNA metabarcoding on faecal samples to genetically characterize zebra nemabiomes for the first time. We generated sequences for several species new to public genetic reference databases, all of which are typical strongyles in wild zebras (i.e., the three species of Cylindropharynx and Cyathostomum montgomeryi), and identified their closest relatives. We also discovered an apparent fungus infecting a quarter of the expelled Crossocephalus viviparus worms, a hyperabundant nematode species in the family Atractidae, hinting at the possibility that zebra host-parasite dynamics may involve a zebra-fungus mutualism. The two zebra species had similar nemabiomes; we found a complete overlap in the list of nematode species they carry and very similar prevalence (i.e., proportion of hosts infected) for the different nematode species. Our study suggests limited host-specificity in zebra strongyles and high potential for transmission between the plains zebra and the endangered Grevy's zebra.
Keywords: DNA metabarcoding; Equid nemabiome; Equid parasitology; Strongyle phylogeny; Zebra nematodes.
© 2021 Published by Elsevier Ltd on behalf of Australian Society for Parasitology.
Conflict of interest statement
None.
Figures



References
-
- Avramenko R.W., Redman E.M., Lewis R., Bichuette M.A., Palmeira B.M., Yazwinski T.A., Gilleard J.S. The use of nemabiome metabarcoding to explore gastro-intestinal nematode species diversity and anthelmintic treatment effectiveness in beef calves. Int. J. Parasitol. 2017;47:893–902. doi: 10.1016/j.ijpara.2017.06.006. - DOI - PubMed
-
- Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. Exploring the gastrointestinal “nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One. 2015;10:1–19. doi: 10.1371/journal.pone.0143559. - DOI - PMC - PubMed
-
- Braga F.R., Carvalho R.O., Araújo J.M., Silva A.R., Araújo J.V., Lima W.S., Tavela A.O., Ferreira S.R. Predatory activity of the fungi Duddingtonia flagrans, Monacrosporium thaumasium, Monacrosporium sinense and Arthrobotrys robusta on Angiostrongylus vasorum first-stage larvae. J. Helminthol. 2009;83:303–308. doi: 10.1017/S0022149X09232342. - DOI - PubMed
-
- Bredtmann C.M., Krücken J., Kuzmina T., Louro M., Madeira de Carvalho L.M., von Samson-Himmelstjerna G. Nuclear and mitochondrial marker sequences reveal close relationship between Coronocyclus coronatus and a potential Cylicostephanus calicatus cryptic species complex. Infect. Genet. Evol. 2019;75:103956. doi: 10.1016/j.meegid.2019.103956. - DOI - PubMed
LinkOut - more resources
Full Text Sources

