In-planta Gene Targeting in Barley Using Cas9 With and Without Geminiviral Replicons
- PMID: 34713258
- PMCID: PMC8525372
- DOI: 10.3389/fgeed.2021.663380
In-planta Gene Targeting in Barley Using Cas9 With and Without Geminiviral Replicons
Abstract
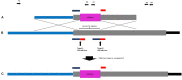
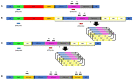
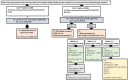
Advances in the use of RNA-guided Cas9-based genome editing in plants have been rapid over the last few years. A desirable application of genome editing is gene targeting (GT), as it allows a wide range of precise modifications; however, this remains inefficient especially in key crop species. Here, we describe successful, heritable gene targeting in barley at the target site of Cas9 using an in-planta strategy but fail to achieve the same using a wheat dwarf virus replicon to increase the copy number of the repair template. Without the replicon, we were able to delete 150 bp of the coding sequence of our target gene whilst simultaneously fusing in-frame mCherry in its place. Starting from 14 original transgenic plants, two plants appeared to have the required gene targeting event. From one of these T0 plants, three independent gene targeting events were identified, two of which were heritable. When the replicon was included, 39 T0 plants were produced and shown to have high copy numbers of the repair template. However, none of the 17 lines screened in T1 gave rise to significant or heritable gene targeting events despite screening twice the number of plants in T1 compared with the non-replicon strategy. Investigation indicated that high copy numbers of repair template created by the replicon approach cause false-positive PCR results which are indistinguishable at the sequence level to true GT events in junction PCR screens widely used in GT studies. In the successful non-replicon approach, heritable gene targeting events were obtained in T1, and subsequently, the T-DNA was found to be linked to the targeted locus. Thus, physical proximity of target and donor sites may be a factor in successful gene targeting.
Keywords: homology-dependent recombination; knock-in; precise insertion; repair template; wheat dwarf virus.
Copyright © 2021 Lawrenson, Hinchliffe, Clarke, Morgan and Harwood.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

