GRP78 facilitates M2 macrophage polarization and tumour progression
- PMID: 34713304
- PMCID: PMC11072571
- DOI: 10.1007/s00018-021-03997-2
GRP78 facilitates M2 macrophage polarization and tumour progression
Abstract
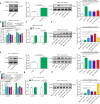
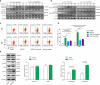
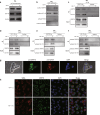
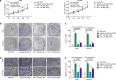
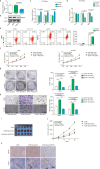
This study investigated the regulation of GRP78 in tumour-associated macrophage polarization in lung cancer. First, our results showed that GRP78 was upregulated in macrophages during M2 polarization and in a conditioned medium derived from lung cancer cells. Next, we found that knocking down GRP78 in macrophages promoted M1 differentiation and suppressed M2 polarization via the Janus kinase/signal transducer and activator of transcription signalling. Moreover, conditioned medium from GRP78- or insulin-like growth factor 1-knockdown macrophages attenuated the survival, proliferation, and migration of lung cancer cells, while conditioned medium from GRP78-overexpressing macrophages had the opposite effects. Additionally, GRP78 knockdown reduced both the secretion of insulin-like growth factor 1 and the phosphorylation of the insulin-like growth factor 1 receptor. Interestingly, insulin-like growth factor 1 neutralization downregulated GRP78 and suppressed GRP78 overexpression-induced M2 polarization. Mechanistically, insulin-like growth factor 1 treatment induced the translocation of GRP78 to the plasma membrane and promoted its association with the insulin-like growth factor 1 receptor. Finally, IGF-1 blockade and knockdown as well as GRP78 knockdown in macrophages inhibited M2 macrophage-induced survival, proliferation, and migration of lung cancer cells both in vitro and in vivo.
Keywords: JAK1/2; NSCLC; STAT3/6; TAM.
© 2021. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, et al. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. - PubMed
-
- Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40(4):558–566. - PubMed
-
- Fuster MM. Targeting the lung cancer microenvironment: harnessing host responses. In: Takiguchi Y, editor. Molecular targeted therapy of lung cancer. Singapore: Springer Singapore; 2017. pp. 309–327.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

