Metagenomic strategies identify diverse integron-integrase and antibiotic resistance genes in the Antarctic environment
- PMID: 34713606
- PMCID: PMC8435808
- DOI: 10.1002/mbo3.1219
Metagenomic strategies identify diverse integron-integrase and antibiotic resistance genes in the Antarctic environment
Abstract
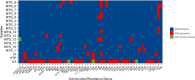
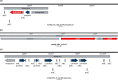
The objective of this study is to identify and analyze integrons and antibiotic resistance genes (ARGs) in samples collected from diverse sites in terrestrial Antarctica. Integrons were studied using two independent methods. One involved the construction and analysis of intI gene amplicon libraries. In addition, we sequenced 17 metagenomes of microbial mats and soil by high-throughput sequencing and analyzed these data using the IntegronFinder program. As expected, the metagenomic analysis allowed for the identification of novel predicted intI integrases and gene cassettes (GCs), which mostly encode unknown functions. However, some intI genes are similar to sequences previously identified by amplicon library analysis in soil samples collected from non-Antarctic sites. ARGs were analyzed in the metagenomes using ABRIcate with CARD database and verified if these genes could be classified as GCs by IntegronFinder. We identified 53 ARGs in 15 metagenomes, but only four were classified as GCs, one in MTG12 metagenome (Continental Antarctica), encoding an aminoglycoside-modifying enzyme (AAC(6´)acetyltransferase) and the other three in CS1 metagenome (Maritime Antarctica). One of these genes encodes a class D β-lactamase (blaOXA-205) and the other two are located in the same contig. One is part of a gene encoding the first 76 amino acids of aminoglycoside adenyltransferase (aadA6), and the other is a qacG2 gene.
Keywords: antibiotic resistance; bioinformatics; horizontal gene transfer; microbial genomics.
© 2021 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Anantharaman, K., Brown, C. T., Hug, L. A., Sharon, I., Castelle, C. J., Probst, A. J., Thomas, B. C., Singh, A., Wilkins, M. J., Karaoz, U., Brodie, E. L., Williams, K. H., Hubbard, S. S., & Banfield, J. F. (2016). Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nature Communications, 7(1), 10.1038/ncomms13219 - DOI - PMC - PubMed
-
- Antelo, V., Guerout, A. M., Mazel, D., Romero, V., Sotelo‐Silveira, J., & Batista, S. (2018). Bacteria from Fildes Peninsula carry class 1 integrons and antibiotic resistance genes in conjugative plasmids. Antarctic Science, 30(1), 22–28. 10.1017/S0954102017000414 - DOI
-
- Antelo, V., Romero, H., & Batista, S. (2015). Detection of integron integrase genes on King George Island, Antarctica. Advance Polar Science, 26, 30–37.
-
- Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., Kubal, M., Meyer, F., Olsen, G. J., Olson, R., Osterman, A. L., Overbeek, R. A., McNeil, L. K., Paarmann, D., Paczian, T., Parrello, B., … Zagnitko, O. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics, 9, 75. 10.1186/1471-2164-9-75 - DOI - PMC - PubMed
-
- Azziz, G., Giménez, M., Romero, H., Valdespino‐Castillo, P. M., Falcón, L. I., Ruberto, L. A. M., Mac Cormack, W. P., & Batista, S. (2019). Detection of presumed genes encoding beta‐lactamases by sequence based screening of metagenomes derived from Antarctic microbial mats. Frontiers of Environmental Science & Engineering, 13, 1–12. 10.1007/s11783-019-1128-1 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

