Status quo of tet regulation in bacteria
- PMID: 34713957
- PMCID: PMC8966031
- DOI: 10.1111/1751-7915.13926
Status quo of tet regulation in bacteria
Abstract
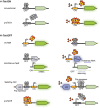
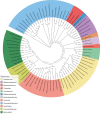
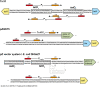
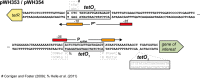
The tetracycline repressor (TetR) belongs to the most popular, versatile and efficient transcriptional regulators used in bacterial genetics. In the tetracycline (Tc) resistance determinant tet(B) of transposon Tn10, tetR regulates the expression of a divergently oriented tetA gene that encodes a Tc antiporter. These components of Tn10 and of other natural or synthetic origins have been used for tetracycline-dependent gene regulation (tet regulation) in at least 40 bacterial genera. Tet regulation serves several purposes such as conditional complementation, depletion of essential genes, modulation of artificial genetic networks, protein overexpression or the control of gene expression within cell culture or animal infection models. Adaptations of the promoters employed have increased tet regulation efficiency and have made this system accessible to taxonomically distant bacteria. Variations of TetR, different effector molecules and mutated DNA binding sites have enabled new modes of gene expression control. This article provides a current overview of tet regulation in bacteria.
© 2021 The Authors. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


References
-
- Agersø, Y. , and Guardabassi, L. (2005) Identification of Tet 39, a novel class of tetracycline resistance determinant in Acinetobacter spp. of environmental and clinical origin. J Antimicrob Chemother 55: 566–569. - PubMed
-
- Armbruster, C.E. , Forsyth‐DeOrnellas, V. , Johnson, A.O. , Smith, S.N. , Zhao, L. , Wu, W. , and Mobley, H.L.T. (2017) Genome‐wide transposon mutagenesis of Proteus mirabilis: essential genes, fitness factors for catheter‐associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13: e1006434. - PMC - PubMed
-
- Baumeister, R. , Helbl, V. , and Hillen, W. (1992) Contacts between Tet repressor and tet operator revealed by new recognition specificities of single amino acid replacement mutants. J Mol Biol 226: 1257–1270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

