Insertion-trigger residues differentially modulate endosomal escape by cytotoxic necrotizing factor toxins
- PMID: 34715130
- PMCID: PMC8592880
- DOI: 10.1016/j.jbc.2021.101347
Insertion-trigger residues differentially modulate endosomal escape by cytotoxic necrotizing factor toxins
Abstract
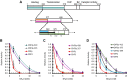
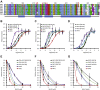
The cellular specificity, potency, and modular nature of bacterial protein toxins enable their application for targeted cytosolic delivery of therapeutic cargo. Efficient endosomal escape is a critical step in the design of bacterial toxin-inspired drug delivery (BTIDD) vehicles to avoid lysosomal degradation and promote optimal cargo delivery. The cytotoxic necrotizing factor (CNF) family of modular toxins represents a useful model for investigating cargo-delivery mechanisms due to the availability of many homologs with high sequence identity, their flexibility in swapping domains, and their differential activity profiles. Previously, we found that CNFy is more sensitive to endosomal acidification inhibitors than CNF1 and CNF2. Here, we report that CNF3 is even less sensitive than CNF1/2. We identified two amino acid residues within the putative translocation domain (E374 and E412 in CNFy, Q373 and S411 in CNF3) that differentiate between these two toxins. Swapping these corresponding residues in each toxin changed the sensitivity to endosomal acidification and efficiency of cargo-delivery to be more similar to the other toxin. Results suggested that trafficking to the more acidic late endosome is required for cargo delivery by CNFy but not CNF3. This model was supported by results from toxin treatment of cells in the presence of NH4Cl, which blocks endosomal acidification, and of small-molecule inhibitors EGA, which blocks trafficking to late endosomes, and ABMA, which blocks endosomal escape and trafficking to the lysosomal degradative pathway. These findings suggest that it is possible to fine-tune endosomal escape and cytosolic cargo delivery efficiency in designing BTIDD platforms.
Keywords: bacterial toxin; drug delivery system; fusion protein; molecular evolution; protein chimera; protein deamidation; protein engineering; protein translocation; small GTPase; structure–function.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Ho M., Chang L.H., Pires-Alves M., Thyagarajan B., Bloom J.E., Gu Z., Aberle K.K., Teymorian S.A., Bannai Y., Johnson S.C., McArdle J.J., Wilson B.A. Recombinant botulinum neurotoxin A heavy chain-based delivery vehicles for neuronal cell targeting. Protein Eng. Des. Sel. 2011;24:247–253. - PMC - PubMed
-
- McNutt P.M., Vazquez-Cintron E.J., Tenezaca L., Ondeck C.A., Kelly K.E., Mangkhalakhili M., Machamer J.B., Angeles C.A., Glotfelty E.J., Cika J., Benjumea C.H., Whitfield J.T., Band P.A., Shoemaker C.B., Ichtchenko K. Neuronal delivery of antibodies has therapeutic effects in animal models of botulism. Sci. Transl. Med. 2021;13 - PMC - PubMed
-
- Miyashita S.I., Zhang J., Zhang S., Shoemaker C.B., Dong M. Delivery of single-domain antibodies into neurons using a chimeric toxin-based platform is therapeutic in mouse models of botulism. Sci. Transl. Med. 2021;13 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

