Over 1000 tools reveal trends in the single-cell RNA-seq analysis landscape
- PMID: 34715899
- PMCID: PMC8555270
- DOI: 10.1186/s13059-021-02519-4
Over 1000 tools reveal trends in the single-cell RNA-seq analysis landscape
Abstract
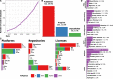
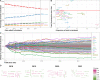
Recent years have seen a revolution in single-cell RNA-sequencing (scRNA-seq) technologies, datasets, and analysis methods. Since 2016, the scRNA-tools database has cataloged software tools for analyzing scRNA-seq data. With the number of tools in the database passing 1000, we provide an update on the state of the project and the field. This data shows the evolution of the field and a change of focus from ordering cells on continuous trajectories to integrating multiple samples and making use of reference datasets. We also find that open science practices reward developers with increased recognition and help accelerate the field.
© 2021. The Author(s).
Conflict of interest statement
FJT reports receiving consulting fees from ImmunAI and an ownership interest in Dermagnostix. The other author declares no competing interests.
Figures

References
-
- Angerer P, Simon L, Tritschler S, Wolf FA, Fischer D, Theis FJ. Single cells make big data: new challenges and opportunities in transcriptomics. Curr Opin Syst Biol. 2017;4:85–91. doi: 10.1016/j.coisb.2017.07.004. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

