Robust Virus-Specific Adaptive Immunity in COVID-19 Patients with SARS-CoV-2 Δ382 Variant Infection
- PMID: 34716845
- PMCID: PMC8556776
- DOI: 10.1007/s10875-021-01142-z
Robust Virus-Specific Adaptive Immunity in COVID-19 Patients with SARS-CoV-2 Δ382 Variant Infection
Erratum in
-
Correction to: Robust Virus-Specific Adaptive Immunity in COVID-19 Patients with SARS-CoV-2 Δ382 Variant Infection.J Clin Immunol. 2022 Feb;42(2):230-231. doi: 10.1007/s10875-021-01184-3. J Clin Immunol. 2022. PMID: 34837161 Free PMC article. No abstract available.
Abstract
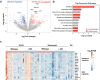
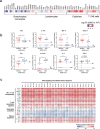
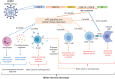
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) that have become dominant as the pandemic progresses bear the ORF8 mutation together with multiple spike mutations. A 382-nucleotide deletion (Δ382) in the ORF7b and ORF8 regions has been associated with milder disease phenotype and less systemic inflammation in COVID-19 patients. However, its impact on host immunity against SARS-CoV-2 remains undefined. Here, RNA-sequencing was performed to elucidate whole blood transcriptomic profiles and identify contrasting immune signatures between patients infected with either wildtype or Δ382 SARS-CoV-2 variant. Interestingly, the immune landscape of Δ382 SARS-CoV-2 infected patients featured an increased adaptive immune response, evidenced by enrichment of genes related to T cell functionality, a more robust SARS-CoV-2-specific T cell immunity, as well as a more rapid antibody response. At the molecular level, eukaryotic initiation factor 2 signaling was found to be upregulated in patients bearing Δ382, and its associated genes were correlated with systemic levels of T cell-associated and pro-inflammatory cytokines. This study provides more in-depth insight into the host-pathogen interactions of ORF8 with great promise as a therapeutic target to combat SARS-CoV-2 infection.
Keywords: Adaptive immune response; Antibody response; CD4+ T cell response; CD8+ T cell response; COVID-19; ORF8; SARS-CoV-2; Transcriptome.
© 2021. The Author(s).
Conflict of interest statement
A patent application for the SFB assay has been filed (Singapore patent 10202009679P: A Method Of Detecting Antibodies And Related Products. YSG, LFPN, and LR). A patent application on the identified linear epitopes S14P5 and S20P2 has also been filed (PCT/SG2021/050178: Antibody-binding linear B cell epitopes of SARS-CoV and SARS-CoV-2. NKWY, SNA, GC, CMP, CYPL, RSLC, LR and LFPN). All other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

