Cis-acting super-enhancer lncRNAs as biomarkers to early-stage breast cancer
- PMID: 34717732
- PMCID: PMC8557595
- DOI: 10.1186/s13058-021-01479-8
Cis-acting super-enhancer lncRNAs as biomarkers to early-stage breast cancer
Abstract
Background: Increased breast cancer screening over the past four decades has led to a substantial rise in the diagnosis of ductal carcinoma in situ (DCIS). Although DCIS lesions precede invasive ductal carcinoma (IDC), they do not always transform into cancer. The current standard-of-care for DCIS is an aggressive course of therapy to prevent invasive and metastatic disease resulting in over-diagnosis and over-treatment. Thus, there is a critical need to identify functional determinants of progression of DCIS to IDC to allow discrimination between indolent and aggressive disease. Recent studies show that super-enhancers, in addition to promoting other gene transcription, are themselves transcribed producing super-enhancer associated long noncoding RNAs (SE-lncRNAs). These SE-lncRNAs can interact with their associated enhancer regions in cis and influence activities and expression of neighboring genes. Furthermore, they represent a novel, untapped group of therapeutic targets.
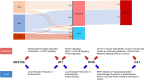
Methods: With an integrative analysis of enhancer loci with global expression of SE-lncRNAs in the MCF10A progression series, we have identified differentially expressed SE-lncRNAs which can identify mechanisms for DCIS to IDC progression. Furthermore, cross-referencing these SE-lncRNAs with patient samples in the The Cancer Genome Atlas (TCGA) database, we have unveiled 27 clinically relevant SE-lncRNAs that potentially interact with their enhancer to regulate nearby gene expression. To complement SE-lncRNA expression studies, we conducted an unbiased global analysis of super-enhancers that are acquired or lost in progression.
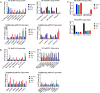
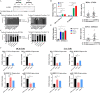
Results: Here we designate SE-lncRNAs RP11-379F4.4 and RP11-465B22.8 as potential markers of progression of DCIS to IDC through regulation of the expression of their neighboring genes (RARRES1 and miR-200b, respectively). Moreover, we classified 403 super-enhancer regions in MCF10A normal cells, 627 in AT1, 1053 in DCIS, and 320 in CA1 cells. Comparison analysis of acquired/lost super-enhancer regions with super-enhancer regions classified in 47 ER positive patients, 10 triple negative breast cancer (TNBC) patients, and 11 TNBC cell lines reveal critically acquired pathways including STAT signaling and NF-kB signaling. In contrast, protein folding, and local estrogen production are identified as major pathways lost in progression.
Conclusion: Collectively, these analyses identify differentially expressed SE-lncRNAs and acquired/lost super-enhancers in progression of breast cancer important for promoting DCIS lesions to IDC.
Keywords: Breast cancer progression; Ductal carcinoma in situ; Super-enhancer long non-coding RNAs; Super-enhancers.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Vaidya JC, Patkar V. Fast facts: early. Breast Cancer. 2016 doi: 10.1159/isbn.978-1-910797-25-9. - DOI
-
- Breast Cancer Facts & Figures. https://www.cancer.org/research/cancer-facts-statistics/breast-cancer-fa.... Accessed 3 Sep 2020
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

