LocalNgsRelate: a software tool for inferring IBD sharing along the genome between pairs of individuals from low-depth NGS data
- PMID: 34718411
- PMCID: PMC8796377
- DOI: 10.1093/bioinformatics/btab732
LocalNgsRelate: a software tool for inferring IBD sharing along the genome between pairs of individuals from low-depth NGS data
Abstract
Motivation: Inference of identity-by-descent (IBD) sharing along the genome between pairs of individuals has important uses. But all existing inference methods are based on genotypes, which is not ideal for low-depth Next Generation Sequencing (NGS) data from which genotypes can only be called with high uncertainty.
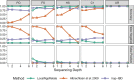
Results: We present a new probabilistic software tool, LocalNgsRelate, for inferring IBD sharing along the genome between pairs of individuals from low-depth NGS data. Its inference is based on genotype likelihoods instead of genotypes, and thereby it takes the uncertainty of the genotype calling into account. Using real data from the 1000 Genomes project, we show that LocalNgsRelate provides more accurate IBD inference for low-depth NGS data than two state-of-the-art genotype-based methods, Albrechtsen et al. (2009) and hap-IBD. We also show that the method works well for NGS data down to a depth of 2×.
Availability and implementation: LocalNgsRelate is freely available at https://github.com/idamoltke/LocalNgsRelate.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press.
Figures