Hsp90 interaction networks in fungi-tools and techniques
- PMID: 34718512
- PMCID: PMC8599792
- DOI: 10.1093/femsyr/foab054
Hsp90 interaction networks in fungi-tools and techniques
Abstract
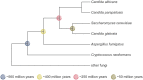
Heat-shock protein 90 (Hsp90) is a central regulator of cellular proteostasis. It stabilizes numerous proteins that are involved in fundamental processes of life, including cell growth, cell-cycle progression and the environmental response. In addition to stabilizing proteins, Hsp90 governs gene expression and controls the release of cryptic genetic variation. Given its central role in evolution and development, it is important to identify proteins and genes that interact with Hsp90. This requires sophisticated genetic and biochemical tools, including extensive mutant collections, suitable epitope tags, proteomics approaches and Hsp90-specific pharmacological inhibitors for chemogenomic screens. These usually only exist in model organisms, such as the yeast Saccharomyces cerevisiae. Yet, the importance of other fungal species, such as Candida albicans and Cryptococcus neoformans, as serious human pathogens accelerated the development of genetic tools to study their virulence and stress response pathways. These tools can also be exploited to map Hsp90 interaction networks. Here, we review tools and techniques for Hsp90 network mapping available in different fungi and provide a summary of existing mapping efforts. Mapping Hsp90 networks in fungal species spanning >500 million years of evolution provides a unique vantage point, allowing tracking of the evolutionary history of eukaryotic Hsp90 networks.
Keywords: Aspergillus fumigatus; Candida; Cryptococcus neoformans; Hsp90; chemogenomics; mutant libraries; protein–protein interactions; proteomics; synthetic lethality.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Figures

Similar articles
-
Global proteomic analyses define an environmentally contingent Hsp90 interactome and reveal chaperone-dependent regulation of stress granule proteins and the R2TP complex in a fungal pathogen.PLoS Biol. 2019 Jul 8;17(7):e3000358. doi: 10.1371/journal.pbio.3000358. eCollection 2019 Jul. PLoS Biol. 2019. PMID: 31283755 Free PMC article.
-
The Hsp90 Chaperone Network Modulates Candida Virulence Traits.Trends Microbiol. 2017 Oct;25(10):809-819. doi: 10.1016/j.tim.2017.05.003. Epub 2017 May 23. Trends Microbiol. 2017. PMID: 28549824 Free PMC article. Review.
-
Adaptation of the tetracycline-repressible system for modulating the expression of essential genes in Cryptococcus neoformans.mSphere. 2025 May 27;10(5):e0101824. doi: 10.1128/msphere.01018-24. Epub 2025 May 1. mSphere. 2025. PMID: 40310102 Free PMC article.
-
Stress-Activated Protein Kinases in Human Fungal Pathogens.Front Cell Infect Microbiol. 2019 Jul 17;9:261. doi: 10.3389/fcimb.2019.00261. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31380304 Free PMC article. Review.
-
Heat shock protein 90 localizes to the surface and augments virulence factors of Cryptococcus neoformans.PLoS Negl Trop Dis. 2017 Aug 4;11(8):e0005836. doi: 10.1371/journal.pntd.0005836. eCollection 2017 Aug. PLoS Negl Trop Dis. 2017. PMID: 28783748 Free PMC article.
Cited by
-
A Preliminary in vitro and in vivo Evaluation of the Effect and Action Mechanism of 17-AAG Combined With Azoles Against Azole-Resistant Candida spp.Front Microbiol. 2022 Jul 7;13:825745. doi: 10.3389/fmicb.2022.825745. eCollection 2022. Front Microbiol. 2022. PMID: 35875545 Free PMC article.
-
Insights and Perspectives on the Role of Proteostasis and Heat Shock Proteins in Fungal Infections.Microorganisms. 2023 Jul 25;11(8):1878. doi: 10.3390/microorganisms11081878. Microorganisms. 2023. PMID: 37630438 Free PMC article. Review.
-
Hsp90: From Cellular to Organismal Proteostasis.Cells. 2022 Aug 10;11(16):2479. doi: 10.3390/cells11162479. Cells. 2022. PMID: 36010556 Free PMC article. Review.
-
Strain variation in gene expression impact of hyphal cyclin Hgc1 in Candida albicans.G3 (Bethesda). 2023 Aug 30;13(9):jkad151. doi: 10.1093/g3journal/jkad151. G3 (Bethesda). 2023. PMID: 37405402 Free PMC article.
-
Retinoids as Alternative Antifungal Agents Against Candida albicans: In Vitro and In Silico Evidence.Microorganisms. 2025 Jan 22;13(2):237. doi: 10.3390/microorganisms13020237. Microorganisms. 2025. PMID: 40005604 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

