VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center
- PMID: 34718728
- PMCID: PMC8728164
- DOI: 10.1093/nar/gkab929
VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center
Abstract
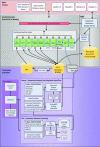
The Eukaryotic Pathogen, Vector and Host Informatics Resource (VEuPathDB, https://veupathdb.org) represents the 2019 merger of VectorBase with the EuPathDB projects. As a Bioinformatics Resource Center funded by the National Institutes of Health, with additional support from the Welllcome Trust, VEuPathDB supports >500 organisms comprising invertebrate vectors, eukaryotic pathogens (protists and fungi) and relevant free-living or non-pathogenic species or hosts. Designed to empower researchers with access to Omics data and bioinformatic analyses, VEuPathDB projects integrate >1700 pre-analysed datasets (and associated metadata) with advanced search capabilities, visualizations, and analysis tools in a graphic interface. Diverse data types are analysed with standardized workflows including an in-house OrthoMCL algorithm for predicting orthology. Comparisons are easily made across datasets, data types and organisms in this unique data mining platform. A new site-wide search facilitates access for both experienced and novice users. Upgraded infrastructure and workflows support numerous updates to the web interface, tools, searches and strategies, and Galaxy workspace where users can privately analyse their own data. Forthcoming upgrades include cloud-ready application architecture, expanded support for the Galaxy workspace, tools for interrogating host-pathogen interactions, and improved interactions with affiliated databases (ClinEpiDB, MicrobiomeDB) and other scientific resources, and increased interoperability with the Bacterial & Viral BRC.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Giraldo-Calderon G.I., Emrich S.J., MacCallum R.M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S., VectorBase C., Madey G. et al. . VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–D713. - PMC - PubMed
-
- Davidson S.B., Crabtree J., Brunk B., Schug J., Tannen V., Overton G.C., Stoeckert J., C. J. K2/Kleisli and GUS: experiments in integrated access to genomic data sources. IBM Syst. J. 2001; 40:512–531.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

