BacDive in 2022: the knowledge base for standardized bacterial and archaeal data
- PMID: 34718743
- PMCID: PMC8728306
- DOI: 10.1093/nar/gkab961
BacDive in 2022: the knowledge base for standardized bacterial and archaeal data
Abstract
The bacterial metadatabase BacDive (https://bacdive.dsmz.de) has developed into a leading database for standardized prokaryotic data on strain level. With its current release (07/2021) the database offers information for 82 892 bacterial and archaeal strains covering taxonomy, morphology, cultivation, metabolism, origin, and sequence information within 1048 data fields. By integrating high-quality data from additional culture collections as well as detailed information from species descriptions, the amount of data provided has increased by 30% over the past three years. A newly developed query builder tool in the advanced search now allows complex database queries. Thereby bacterial strains can be systematically searched based on combinations of their attributes, e.g. growth and metabolic features for biotechnological applications or to identify gaps in the present knowledge about bacteria. A new interactive dashboard provides a statistic overview over the most important data fields. Additional new features are improved genomic sequence data, integrated NCBI TaxIDs and links to BacMedia, the new sister database on cultivation media. To improve the findability and interpretation of data through search engines, data in BacDive are annotated with bioschemas.org terms.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
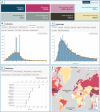
References
-
- Overmann J., Abt B., Sikorski J.. Present and future of culturing bacteria. Annu. Rev. Microbiol. 2017; 71:711–730. - PubMed
-
- Brinker T.J., Hekler A., Enk A.H., Klode J., Hausschild A., Berking C., Schilling B., Haferkamp S., Schadendorf D., Holland-Letz T.et al. .. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer. 2019; 113:47–54. - PubMed
-
- Li G., Rabe K.S., Nielsen J., Engqvist M.K.M.. Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. ACS Synth. Biol. 2019; 8:1411–1420. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

