Misregulation of MYB16 expression causes stomatal cluster formation by disrupting polarity during asymmetric cell divisions
- PMID: 34718767
- PMCID: PMC8774054
- DOI: 10.1093/plcell/koab260
Misregulation of MYB16 expression causes stomatal cluster formation by disrupting polarity during asymmetric cell divisions
Abstract
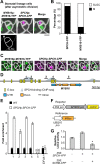
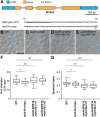
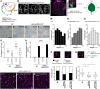
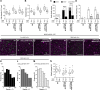
Stomatal pores and the leaf cuticle regulate evaporation from the plant body and balance the tradeoff between photosynthesis and water loss. MYB16, encoding a transcription factor involved in cutin biosynthesis, is expressed in stomatal lineage ground cells, suggesting a link between cutin biosynthesis and stomatal development. Here, we show that the downregulation of MYB16 in meristemoids is directly mediated by the stomatal master transcription factor SPEECHLESS (SPCH) in Arabidopsis thaliana. The suppression of MYB16 before an asymmetric division is crucial for stomatal patterning, as its overexpression or ectopic expression in meristemoids increased stomatal density and resulted in the formation of stomatal clusters, as well as affecting the outer cell wall structure. Expressing a cutinase gene in plants ectopically expressing MYB16 reduced stomatal clustering, suggesting that cutin affects stomatal signaling or the polarity setup in asymmetrically dividing cells. The clustered stomatal phenotype was rescued by overexpressing EPIDERMAL PATTERNING FACTOR2, suggesting that stomatal signaling was still functional in these plants. Growing seedlings ectopically expressing MYB16 on high-percentage agar plates to modulate tensile strength rescued the polarity and stomatal cluster defects of these seedlings. Therefore, the inhibition of MYB16 expression by SPCH in the early stomatal lineage is required to correctly place the polarity protein needed for stomatal patterning during leaf morphogenesis.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




Comment in
-
Back to the roots: A focus on plant cell biology.Plant Cell. 2022 Jan 20;34(1):1-3. doi: 10.1093/plcell/koab278. Plant Cell. 2022. PMID: 34755878 Free PMC article. No abstract available.
-
MYB16 expression in the stomatal lineage: Wrong place at the wrong time leads to stomata side-by-side.Plant Cell. 2022 Jan 20;34(1):8-9. doi: 10.1093/plcell/koab268. Plant Cell. 2022. PMID: 35226743 Free PMC article. No abstract available.
References
-
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 - PubMed
-
- Bessire M, Borel S, Fabre G, Carrac L, Efremova N, Yephremov A, Cao Y, Jetter R, Jacquat AC, Métraux JP, et al. (2011). A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970 - PMC - PubMed
-
- Bhanot V, Fadanavis SV, Panwar J (2021) Revisiting the architecture, biosynthesis and functional aspects of the plant cuticle: there is more scope. Environ Exp Bot 183: 104364
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

