Genetic analysis of single disseminated tumor cells in the lymph nodes and bone marrow of patients with head and neck squamous cell carcinoma
- PMID: 34719102
- PMCID: PMC8763651
- DOI: 10.1002/1878-0261.13113
Genetic analysis of single disseminated tumor cells in the lymph nodes and bone marrow of patients with head and neck squamous cell carcinoma
Abstract
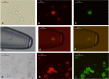
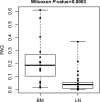
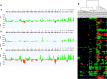
Considering the limited information on the biology and molecular characteristics of disseminated tumor cells (DTCs) in head and neck squamous cell carcinoma (HNSCC), we examined the genomic alterations in DTCs from HNSCCs and their potential clinical relevance. To analyze both the lymphatic and hematogenous routes of tumor cell dissemination, we investigated samples from lymph nodes (LNs) and bone marrow (BM) of 49 patients using immunofluorescence double staining for epithelial cells expressing cytokeratin 18 (KRT18) and/or epithelial cell adhesion molecules (EpCAM, CD326). The identified marker-positive cells were isolated by micromanipulation followed by single-cell whole-genome amplification and metaphase-based comparative genomic hybridization (mCGH) to determine genome-wide copy number alterations. The findings were correlated with clinical parameters and follow-up data. We detected chromosomal aberrations in KRT18- and EpCAM-positive cells from both compartments; BM-derived cells showed a significantly higher percentage of aberrant genome (PAG) per cell than cells detected in LNs. No significant association was found between DTC data and clinical follow-up. Genomic profiling of BM-DTCs revealed genomic alterations typical for HNSCC, suggesting hematogenous dissemination of subclones around the time of surgery. In contrast, DTC data in LNs revealed that several marker-positive cells were not of malignant origin, indicating the presence of epithelial glandular inclusions in parts of the processed neck LN samples. Therefore, DTC detection of LNs in the neck based only on epithelial markers is not advisable and requires detection of chromosomal instability (CIN), gene mutations, or additional markers, which have yet to be identified. Nevertheless, our investigation paves the way for larger studies to focus on HNSCC BM-DTCs with high-resolution methods to gain deeper insights into the biology of hematogenous metastasis in this cancer.
Keywords: bone marrow; disseminated tumor cells; genetic alterations; head and neck squamous cell carcinoma; lymph nodes; minimal residual disease.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Disseminated tumour cells with highly aberrant genomes are linked to poor prognosis in operable oesophageal adenocarcinoma.Br J Cancer. 2017 Aug 22;117(5):725-733. doi: 10.1038/bjc.2017.233. Epub 2017 Jul 20. Br J Cancer. 2017. PMID: 28728164 Free PMC article.
-
Immunohistochemical detection of lymph node-DTCs in patients with node-negative HNSCC.Int J Cancer. 2017 May 1;140(9):2112-2124. doi: 10.1002/ijc.30617. Epub 2017 Feb 8. Int J Cancer. 2017. PMID: 28120418
-
Molecular Markers of Occult Lymph Node Metastasis in Head and Neck Squamous Cell Carcinoma (HNSCC) Patients.Front Biosci (Landmark Ed). 2025 Feb 20;30(2):25267. doi: 10.31083/FBL25267. Front Biosci (Landmark Ed). 2025. PMID: 40018925 Review.
-
Bone marrow versus sentinel lymph node involvement in breast cancer: a comparison of early hematogenous and early lymphatic tumor spread.Breast Cancer Res Treat. 2012 Jan;131(2):501-8. doi: 10.1007/s10549-011-1802-x. Epub 2011 Oct 5. Breast Cancer Res Treat. 2012. PMID: 21971730
-
Minimal Residual Disease in Head and Neck Cancer and Esophageal Cancer.Adv Exp Med Biol. 2018;1100:55-82. doi: 10.1007/978-3-319-97746-1_4. Adv Exp Med Biol. 2018. PMID: 30411260 Review.
Cited by
-
Single-Cell Molecular Profiling of Head and Neck Squamous Cell Carcinoma Reveals Five Dysregulated Signaling Pathways Associated With Circulating Tumor Cells.Cancer Control. 2024 Jan-Dec;31:10732748241251571. doi: 10.1177/10732748241251571. Cancer Control. 2024. PMID: 38869038 Free PMC article.
-
Temporal and spatial characteristics of tumor evolution in a mouse model of oral squamous cell carcinoma.BMC Cancer. 2022 Nov 24;22(1):1209. doi: 10.1186/s12885-022-10256-5. BMC Cancer. 2022. PMID: 36424557 Free PMC article.
-
Exploring the frontiers: tumor immune microenvironment and immunotherapy in head and neck squamous cell carcinoma.Discov Oncol. 2024 Jan 31;15(1):22. doi: 10.1007/s12672-024-00870-z. Discov Oncol. 2024. PMID: 38294629 Free PMC article. Review.
-
Molecular classification of human papilloma virus-negative head and neck squamous cell carcinomas: Cell cycle-based classifier and prognostic signature.PLoS One. 2023 Oct 30;18(10):e0286414. doi: 10.1371/journal.pone.0286414. eCollection 2023. PLoS One. 2023. PMID: 37903125 Free PMC article.
References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E & Forman D (2011) Global cancer statistics. CA Cancer J Clin 61, 69–90. - PubMed
-
- Chin D, Boyle GM, Porceddu S, Theile DR, Parsons PG & Coman WB (2006) Head and neck cancer: past, present and future. Expert Rev Anticancer Ther 6, 1111–1118. - PubMed
-
- Gath HJ & Brakenhoff RH (1999) Minimal residual disease in head and neck cancer. Cancer Metastasis Rev 18, 109–126. - PubMed
-
- Cramer JD, Burtness B, Le QT & Ferris RL (2019) The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol 16, 669–683. - PubMed
-
- Kulasinghe A, Perry C, Jovanovic L, Nelson C & Punyadeera C (2015) Circulating tumour cells in metastatic head and neck cancers. Int J Cancer 136, 2515–2523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

