Apolipoprotein D as a Potential Biomarker and Construction of a Transcriptional Regulatory-Immune Network Associated with Osteoarthritis by Weighted Gene Coexpression Network Analysis
- PMID: 34719950
- PMCID: PMC8808834
- DOI: 10.1177/19476035211053824
Apolipoprotein D as a Potential Biomarker and Construction of a Transcriptional Regulatory-Immune Network Associated with Osteoarthritis by Weighted Gene Coexpression Network Analysis
Abstract
Objective: Synovial inflammation influences the progression of osteoarthritis (OA). Herein, we aimed to identify potential biomarkers and analyze transcriptional regulatory-immune mechanism of synovitis in OA using weighted gene coexpression network analysis (WGCNA).
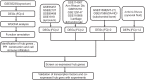
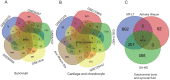
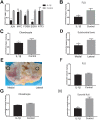
Design: A data set of OA synovium samples (GSE55235) was analyzed based on WGCNA. The most significant module with OA was identified and function annotation of the module was performed, following which the hub genes of the module were identified using Pearson correlation and a protein-protein interaction network was constructed. A transcriptional regulatory network of hub genes was constructed using the TRRUST database. The immune cell infiltration of OA samples was evaluated using the single-sample Gene Set Enrichment Analysis (ssGSEA) method. The hub genes coexpressed in multiple tissues were then screened out using data sets of synovium, cartilage, chondrocyte, subchondral bone, and synovial fluid samples. Finally, transcriptional factors and coexpressed hub genes were validated via experiments.
Results: The turquoise module of GSE55235 was identified via WGCNA. Functional annotation analysis showed that "mineral absorption" and "FoxO signaling pathway" were mostly enriched in the module. JUN, EGR1, FOSB, and KLF4 acted as central nodes in protein-protein interaction network and transcription factors to connect several target genes. "Activated B cell," "activated CD4T cell," "eosinophil," "neutrophil," and "type 17 T helper cell" showed high immune infiltration, while FOSB, KLF6, and MYBL2 showed significant negative correlation with type 17 T helper cell.
Conclusions: Our results suggest that the expression level of apolipoprotein D (APOD) was correlated with OA. Furthermore, transcriptional regulatory-immune network was constructed, which may contribute to OA therapy.
Keywords: hub genes; multiple tissues; osteoarthritis; transcriptional regulatory-immune network; weighted gene coexpression network analysis.
Conflict of interest statement
Figures






References
-
- Postler A, Goronzy J, Günther KP, Lange T, Redeker I, Schmitt J, et al.. Which disease-related factors influence patients’ and physicians’ willingness to consider joint replacement in hip and knee OA? Results of a questionnaire survey linked to claims data. BMC Musculoskelet Disord. 2020;21:352. doi:10.1186/s12891-020-03368-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

