DEER and RIDME Measurements of the Nitroxide-Spin Labelled Copper-Bound Amine Oxidase Homodimer from Arthrobacter Globiformis
- PMID: 34720439
- PMCID: PMC8550341
- DOI: 10.1007/s00723-021-01321-6
DEER and RIDME Measurements of the Nitroxide-Spin Labelled Copper-Bound Amine Oxidase Homodimer from Arthrobacter Globiformis
Abstract
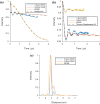
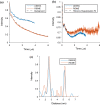
In the study of biological structures, pulse dipolar spectroscopy (PDS) is used to elucidate spin-spin distances at nanometre-scale by measuring dipole-dipole interactions between paramagnetic centres. The PDS methods of Double Electron Electron Resonance (DEER) and Relaxation Induced Dipolar Modulation Enhancement (RIDME) are employed, and their results compared, for the measurement of the dipolar coupling between nitroxide spin labels and copper-II (Cu(II)) paramagnetic centres within the copper amine oxidase from Arthrobacter globiformis (AGAO). The distance distribution results obtained indicate that two distinct distances can be measured, with the longer of these at c.a. 5 nm. Conditions for optimising the RIDME experiment such that it may outperform DEER for these long distances are discussed. Modelling methods are used to show that the distances obtained after data analysis are consistent with the structure of AGAO.
Supplementary information: The online version contains supplementary material available at 10.1007/s00723-021-01321-6.
© The Author(s) 2021.
Figures



References
-
- D. Goldfarb and S. Stoll, EPR Spectroscopy: Fundamentals and Methods. 2018: Wiley.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
