The Use of Artificial Neural Networks to Forecast the Behavior of Agent-Based Models of Pathophysiology: An Example Utilizing an Agent-Based Model of Sepsis
- PMID: 34721057
- PMCID: PMC8552109
- DOI: 10.3389/fphys.2021.716434
The Use of Artificial Neural Networks to Forecast the Behavior of Agent-Based Models of Pathophysiology: An Example Utilizing an Agent-Based Model of Sepsis
Abstract
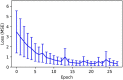
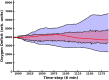
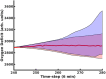
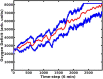
Introduction: Disease states are being characterized at finer and finer levels of resolution via biomarker or gene expression profiles, while at the same time. Machine learning (ML) is increasingly used to analyze and potentially classify or predict the behavior of biological systems based on such characterization. As ML applications are extremely data-intensive, given the relative sparsity of biomedical data sets ML training of artificial neural networks (ANNs) often require the use of synthetic training data. Agent-based models (ABMs) that incorporate known biological mechanisms and their associated stochastic properties are a potential means of generating synthetic data. Herein we present an example of ML used to train an artificial neural network (ANN) as a surrogate system used to predict the time evolution of an ABM focusing on the clinical condition of sepsis. Methods: The disease trajectories for clinical sepsis, in terms of temporal cytokine and phenotypic dynamics, can be interpreted as a random dynamical system. The Innate Immune Response Agent-based Model (IIRABM) is a well-established model that utilizes known cellular and molecular rules to simulate disease trajectories corresponding to clinical sepsis. We have utilized two distinct neural network architectures, Long Short-Term Memory and Multi-Layer Perceptron, to take a time sequence of five measurements of eleven IIRABM simulated serum cytokine concentrations as input and to return both the future cytokine trajectories as well as an aggregate metric representing the patient's state of health. Results: The ANNs predicted model trajectories with the expected amount of error, due to stochasticity in the simulation, and recognizing that the mapping from a specific cytokine profile to a state-of-health is not unique. The Multi-Layer Perceptron neural network, generated predictions with a more accurate forecasted trajectory cone. Discussion: This work serves as a proof-of-concept for the use of ANNs to predict disease progression in sepsis as represented by an ABM. The findings demonstrate that multicellular systems with intrinsic stochasticity can be approximated with an ANN, but that forecasting a specific trajectory of the system requires sequential updating of the system state to provide a rolling forecast horizon.
Keywords: agent-based model (ABM); machine learning; neural networks; sepsis; time series.
Copyright © 2021 Larie, An and Cockrell.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

