B Cell Mobilization, Dissemination, Fine Tuning of Local Antigen Specificity and Isotype Selection in Asthma
- PMID: 34721376
- PMCID: PMC8552043
- DOI: 10.3389/fimmu.2021.702074
B Cell Mobilization, Dissemination, Fine Tuning of Local Antigen Specificity and Isotype Selection in Asthma
Abstract
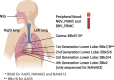
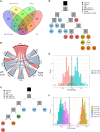
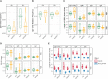
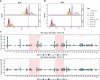
In order to better understand how the immune system interacts with environmental triggers to produce organ-specific disease, we here address the hypothesis that B and plasma cells are free to migrate through the mucosal surfaces of the upper and lower respiratory tracts, and that their total antibody repertoire is modified in a common respiratory tract disease, in this case atopic asthma. Using Adaptive Immune Receptor Repertoire sequencing (AIRR-seq) we have catalogued the antibody repertoires of B cell clones retrieved near contemporaneously from multiple sites in the upper and lower respiratory tract mucosa of adult volunteers with atopic asthma and non-atopic controls and traced their migration. We show that the lower and upper respiratory tracts are immunologically connected, with trafficking of B cells directionally biased from the upper to the lower respiratory tract and points of selection when migrating from the nasal mucosa and into the bronchial mucosa. The repertoires are characterized by both IgD-only B cells and others undergoing class switch recombination, with restriction of the antibody repertoire distinct in asthmatics compared with controls. We conclude that B cells and plasma cells migrate freely throughout the respiratory tract and exhibit distinct antibody repertoires in health and disease.
Keywords: IgD; adaptive immunity; allergy; asthma; immunoglobulin; innate immunity; repertoire and AIRR-seq/sequencing.
Copyright © 2021 Ohm-Laursen, Meng, Hoehn, Nouri, Jiang, Clouser, Johnstone, Hause, Sandhar, Upton, Chevretton, Lakhani, Corrigan, Kleinstein and Gould.
Conflict of interest statement
SHK receives consulting fees from Northrop Grumman. KBH receives consulting fees from Prellis Biologics. YJ, CC, TGJ and RH are employed by Bristol Myers Squibb. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor has declared past collaborations with one of the authors SHK at the time of this review.
Figures