Pumpkin (Cucurbita moschata) HSP20 Gene Family Identification and Expression Under Heat Stress
- PMID: 34721541
- PMCID: PMC8553033
- DOI: 10.3389/fgene.2021.753953
Pumpkin (Cucurbita moschata) HSP20 Gene Family Identification and Expression Under Heat Stress
Abstract
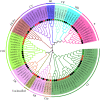
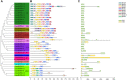
Pumpkin (Cucurbita moschata) is an important cucurbit vegetable crop that has strong resistance to abiotic stress. While heat shock protein 20 (HSP20) has been implicated in vegetable response to heat stress, little is known regarding activity of HSP20 family proteins in C. moschata. Here, we performed a comprehensive genome-wide analysis to identify and characterize the functional dynamics of the Cucurbita moschata HSP20 (CmoHSP20) gene family. A total of 33 HSP20 genes distributed across 13 chromosomes were identified from the pumpkin genome. Our phylogenetic analysis determined that the CmoHSP20 proteins fell into nine distinct subfamilies, a division supported by the conserved motif composition and gene structure analyses. Segmental duplication events were shown to play a key role in expansion of the CmoHSP20 gene family. Synteny analysis revealed that 19 and 18 CmoHSP20 genes were collinear with those in the cucumber and melon genomes, respectively. Furthermore, the expression levels of pumpkin HSP20 genes were differentially induced by heat stress. The transcript level of CmoHSP20-16, 24 and 25 were down-regulated by heat stress, while CmoHSP20-7, 13, 18, 22, 26 and 32 were up-regulated by heat stress, which could be used as heat tolerance candidate genes. Overall, these findings contribute to our understanding of vegetable HSP20 family genes and provide valuable information that can be used to breed heat stress resistance in cucurbit vegetable crops.
Keywords: Cucurbita moschata; gene expression; gene family; heat shock protein 20; heat stress.
Copyright © 2021 Hu, Zhang, Liu, Li, Wang, Zhu, Liao, Yun, Huang, Zhang and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Aghdam M. S., Sevillano L., Flores F. B., Bodbodak S. (2013). Heat Shock Proteins as Biochemical Markers for Postharvest Chilling Stress in Fruits and Vegetables. Scientia Horticult. 160, 54–64. 10.1016/j.scienta.2013.05.020 - DOI
LinkOut - more resources
Full Text Sources

