Characterization of peptide binding to the SARS-CoV-2 host factor neuropilin
- PMID: 34722943
- PMCID: PMC8540010
- DOI: 10.1016/j.heliyon.2021.e08251
Characterization of peptide binding to the SARS-CoV-2 host factor neuropilin
Abstract
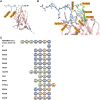
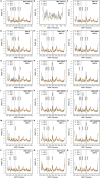
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a global health concern. It is now well established that the spike (S) protein of SARS-CoV-2 interacts with its primary host receptor, the angiotensin converting enzyme 2 (ACE2). Additionally, the interaction of S with the neuropilin (NRP) receptor has been reported to facilitate viral entry. SARS-CoV-2 S protein binds to neuropilin-1 (NRP1) by virtue of a CendR motif which terminates with either an arginine or lysine. Furthermore, a number of different peptide sequences have been reported to bind to the same site in NRP1 including vascular endothelial growth factor A and other viral proteins. To gain a deeper understanding of additional factors besides the C-terminal arginine that may favour high NRP1 binding, several modelled peptides were investigated using triplicate 1 μs molecular dynamics simulations. A C-end histidine failed to exhibit strong NRP1 affinity. Some previously reported factors that increase binding affinity and secure NRP1 receptor activation was observed in the NRP1-peptide complexes studied and such complexes had higher molecular mechanics-generalized Born surface area based free energy of binding. Additionally, the results also highlight the relevance of an exposed arginine at its canonical location as capping it blocked arginine from engaging key residues at the NRP1 receptor site that are indispensable for functional binding; and that the presence of proline reinforces the C-terminal arginine. Given that stable NRP1 binding is crucial for viral uptake, stable interactions should be accounted for in the design of potential drugs and treatment routes to target or disrupt this interface, considering the S1-NRP1 interaction as well as its endogenous VEGF-A ligand that is associated with nociception.
Keywords: CendR motif; Molecular dynamics; Neuropilin; Spike protein; VEGF-A.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Molecular basis of the new COVID-19 target neuropilin-1 in complex with SARS-CoV-2 S1 C-end rule peptide and small-molecule antagonists.J Mol Liq. 2021 Aug 1;335:116537. doi: 10.1016/j.molliq.2021.116537. Epub 2021 May 20. J Mol Liq. 2021. PMID: 34031621 Free PMC article.
-
Neuropilins: C-end rule peptides and their association with nociception and COVID-19.Comput Struct Biotechnol J. 2021;19:1889-1895. doi: 10.1016/j.csbj.2021.03.025. Epub 2021 Mar 26. Comput Struct Biotechnol J. 2021. PMID: 33815686 Free PMC article. Review.
-
Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of Spike protein S1 and S2.Biophys J. 2021 Jul 20;120(14):2828-2837. doi: 10.1016/j.bpj.2021.05.026. Epub 2021 Jun 2. Biophys J. 2021. PMID: 34087218 Free PMC article.
-
Discovery of natural products to block SARS-CoV-2 S-protein interaction with Neuropilin-1 receptor: A molecular dynamics simulation approach.Microb Pathog. 2022 Sep;170:105701. doi: 10.1016/j.micpath.2022.105701. Epub 2022 Aug 10. Microb Pathog. 2022. PMID: 35963279 Free PMC article.
-
Neuropilin 1: A Novel Entry Factor for SARS-CoV-2 Infection and a Potential Therapeutic Target.Biologics. 2021 May 6;15:143-152. doi: 10.2147/BTT.S307352. eCollection 2021. Biologics. 2021. PMID: 33986591 Free PMC article. Review.
Cited by
-
Mutating novel interaction sites in NRP1 reduces SARS-CoV-2 spike protein internalization.iScience. 2023 Apr 21;26(4):106274. doi: 10.1016/j.isci.2023.106274. Epub 2023 Feb 25. iScience. 2023. PMID: 36910328 Free PMC article.
-
Sars-Cov2 Induced Biochemical Mechanisms in Liver Damage and Intestinal Lesions.Indian J Clin Biochem. 2022 Nov 12;38(4):1-10. doi: 10.1007/s12291-022-01089-x. Online ahead of print. Indian J Clin Biochem. 2022. PMID: 36407686 Free PMC article.
References
-
- Baek D.-S., Kim J.-H., Kim Y.-J., Kim Y.-S. Immunoglobulin Fc-fused peptide without C-terminal Arg or Lys residue augments neuropilin-1-dependent tumor vascular permeability. Mol. Pharm. 2018;15:394–402. - PubMed
-
- Bowers K. SC ’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, Florida. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. 43–43.
LinkOut - more resources
Full Text Sources
Miscellaneous

