DNA Viral Diversity, Abundance, and Functional Potential Vary across Grassland Soils with a Range of Historical Moisture Regimes
- PMID: 34724822
- PMCID: PMC8567247
- DOI: 10.1128/mBio.02595-21
DNA Viral Diversity, Abundance, and Functional Potential Vary across Grassland Soils with a Range of Historical Moisture Regimes
Abstract
Soil viruses are abundant, but the influence of the environment and climate on soil viruses remains poorly understood. Here, we addressed this gap by comparing the diversity, abundance, lifestyle, and metabolic potential of DNA viruses in three grassland soils with historical differences in average annual precipitation, low in eastern Washington (WA), high in Iowa (IA), and intermediate in Kansas (KS). Bioinformatics analyses were applied to identify a total of 2,631 viral contigs, including 14 complete viral genomes from three deep metagenomes (1 terabase [Tb] each) that were sequenced from bulk soil DNA. An additional three replicate metagenomes (∼0.5 Tb each) were obtained from each location for statistical comparisons. Identified viruses were primarily bacteriophages targeting dominant bacterial taxa. Both viral and host diversity were higher in soil with lower precipitation. Viral abundance was also significantly higher in the arid WA location than in IA and KS. More lysogenic markers and fewer clustered regularly interspaced short palindromic repeats (CRISPR) spacer hits were found in WA, reflecting more lysogeny in historically drier soil. More putative auxiliary metabolic genes (AMGs) were also detected in WA than in the historically wetter locations. The AMGs occurring in 18 pathways could potentially contribute to carbon metabolism and energy acquisition in their hosts. Structural equation modeling (SEM) suggested that historical precipitation influenced viral life cycle and selection of AMGs. The observed and predicted relationships between soil viruses and various biotic and abiotic variables have value for predicting viral responses to environmental change. IMPORTANCE Soil viruses are abundant but poorly understood. Because soil viruses regulate the dynamics of their hosts and potentially key processes in soil ecology, it is important to understand them better. Here, we leveraged massive DNA sequencing to unearth previously unknown soil viruses. We found that soil viruses differed across a historical gradient of precipitation. We compared soil viruses from Iowa, which is traditionally wetter, to those from Washington, which is traditionally drier, and from Kansas, which is intermediate. This study provides strong evidence that changes in historical precipitation impact not only the types of soil viruses but also their functional potential.
Keywords: auxiliary metabolic gene; grassland soil; lysogeny; metagenome; soil bacteriophage; soil virus.
Figures


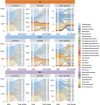
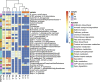

References
-
- Emerson JB, Roux S, Brum JR, Bolduc B, Woodcroft BJ, Jang HB, Singleton CM, Solden LM, Naas AE, Boyd JA, Hodgkins SB, Wilson RM, Trubl G, Li C, Frolking S, Pope PB, Wrighton KC, Crill PM, Chanton JP, Saleska SR, Tyson GW, Rich VI, Sullivan MB. 2018. Host-linked soil viral ecology along a permafrost thaw gradient. Nat Microbiol 3:870–880. doi:10.1038/s41564-018-0190-y. - DOI - PMC - PubMed
-
- Trubl G, Jang HB, Roux S, Emerson JB, Solonenko N, Vik DR, Solden L, Ellenbogen J, Runyon AT, Bolduc B, Woodcroft BJ, Saleska SR, Tyson GW, Wrighton KC, Sullivan MB, Rich VI. 2018. Soil viruses are underexplored players in ecosystem carbon processing. mSystems 3:e00076-18. doi:10.1128/mSystems.00076-18. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
