Cryo-EM structures of PI3Kα reveal conformational changes during inhibition and activation
- PMID: 34725156
- PMCID: PMC8609346
- DOI: 10.1073/pnas.2109327118
Cryo-EM structures of PI3Kα reveal conformational changes during inhibition and activation
Abstract
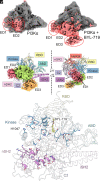
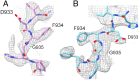
Phosphoinositide 3-kinases (PI3Ks) are lipid kinases essential for growth and metabolism. Their aberrant activation is associated with many types of cancers. Here we used single-particle cryoelectron microscopy (cryo-EM) to determine three distinct conformations of full-length PI3Kα (p110α-p85α): the unliganded heterodimer PI3Kα, PI3Kα bound to the p110α-specific inhibitor BYL-719, and PI3Kα exposed to an activating phosphopeptide. The cryo-EM structures of unbound and of BYL-719-bound PI3Kα are in general accord with published crystal structures. Local deviations are presented and discussed. BYL-719 stabilizes the structure of PI3Kα, but three regions of low-resolution extra density remain and are provisionally assigned to the cSH2, BH, and SH3 domains of p85. One of the extra density regions is in contact with the kinase domain blocking access to the catalytic site. This conformational change indicates that the effects of BYL-719 on PI3Kα activity extend beyond competition with adenosine triphosphate (ATP). In unliganded PI3Kα, the DFG motif occurs in the "in" and "out" positions. In BYL-719-bound PI3Kα, only the DFG-in position, corresponding to the active conformation of the kinase, was observed. The phosphopeptide-bound structure of PI3Kα is composed of a stable core resolved at 3.8 Å. It contains all p110α domains except the adaptor-binding domain (ABD). The p85α domains, linked to the core through the ABD, are no longer resolved, implying that the phosphopeptide activates PI3Kα by fully releasing the niSH2 domain from binding to p110α. The structures presented here show the basal form of the full-length PI3Kα dimer and document conformational changes related to the activated and inhibited states.
Keywords: activation; activity-dependent conformational changes; inhibition; phosphoinositide 3-kinase (PI3K).
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3Kα.Proc Natl Acad Sci U S A. 2022 Nov 16;119(46):e2215621119. doi: 10.1073/pnas.2215621119. Epub 2022 Nov 7. Proc Natl Acad Sci U S A. 2022. PMID: 36343266 Free PMC article.
-
Structural and mechanistic insights provided by single particle cryo-EM analysis of phosphoinositide 3-kinase (PI3Kα).Biochim Biophys Acta Rev Cancer. 2023 Sep;1878(5):188947. doi: 10.1016/j.bbcan.2023.188947. Epub 2023 Jun 30. Biochim Biophys Acta Rev Cancer. 2023. PMID: 37394020 Free PMC article. Review.
-
Nanobodies and chemical cross-links advance the structural and functional analysis of PI3Kα.Proc Natl Acad Sci U S A. 2022 Sep 20;119(38):e2210769119. doi: 10.1073/pnas.2210769119. Epub 2022 Sep 12. Proc Natl Acad Sci U S A. 2022. PMID: 36095215 Free PMC article.
-
Regulation of lipid binding underlies the activation mechanism of class IA PI3-kinases.Oncogene. 2012 Aug 9;31(32):3655-66. doi: 10.1038/onc.2011.532. Epub 2011 Nov 28. Oncogene. 2012. PMID: 22120714 Free PMC article.
-
Molecular Mechanisms of Human Disease Mediated by Oncogenic and Primary Immunodeficiency Mutations in Class IA Phosphoinositide 3-Kinases.Front Immunol. 2018 Mar 19;9:575. doi: 10.3389/fimmu.2018.00575. eCollection 2018. Front Immunol. 2018. PMID: 29616047 Free PMC article. Review.
Cited by
-
Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3Kα.Proc Natl Acad Sci U S A. 2022 Nov 16;119(46):e2215621119. doi: 10.1073/pnas.2215621119. Epub 2022 Nov 7. Proc Natl Acad Sci U S A. 2022. PMID: 36343266 Free PMC article.
-
Functional selection in SH3-mediated activation of the PI3 kinase.bioRxiv [Preprint]. 2024 Apr 30:2024.04.30.591319. doi: 10.1101/2024.04.30.591319. bioRxiv. 2024. PMID: 38746413 Free PMC article. Preprint.
-
Structural insights into isoform-specific RAS-PI3Kα interactions and the role of RAS in PI3Kα activation.Nat Commun. 2025 Jan 9;16(1):525. doi: 10.1038/s41467-024-55766-x. Nat Commun. 2025. PMID: 39788953 Free PMC article.
-
The orchestrated signaling by PI3Kα and PTEN at the membrane interface.Comput Struct Biotechnol J. 2022 Oct 7;20:5607-5621. doi: 10.1016/j.csbj.2022.10.007. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36284707 Free PMC article. Review.
-
Unveiling molecular insights into the mechanism of activation of oncogenic phosphoinositide 3-kinase mutants.Proc Natl Acad Sci U S A. 2022 Dec 13;119(50):e2218237119. doi: 10.1073/pnas.2218237119. Epub 2022 Dec 7. Proc Natl Acad Sci U S A. 2022. PMID: 36475945 Free PMC article. No abstract available.
References
-
- Vanhaesebroeck B., Leevers S. J., Panayotou G., Waterfield M. D., Phosphoinositide 3-kinases: A conserved family of signal transducers. Trends Biochem. Sci. 22, 267–272 (1997). - PubMed
-
- Rameh L. E., Cantley L. C., The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 274, 8347–8350 (1999). - PubMed
-
- Katso R., et al. ., Cellular function of phosphoinositide 3-kinases: Implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 17, 615–675 (2001). - PubMed
-
- Fry M. J., Structure, regulation and function of phosphoinositide 3-kinases. Biochim. Biophys. Acta 1226, 237–268 (1994). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

