PERK signaling through C/EBPδ contributes to ER stress-induced expression of immunomodulatory and tumor promoting chemokines by cancer cells
- PMID: 34725321
- PMCID: PMC8560861
- DOI: 10.1038/s41419-021-04318-y
PERK signaling through C/EBPδ contributes to ER stress-induced expression of immunomodulatory and tumor promoting chemokines by cancer cells
Abstract
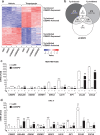
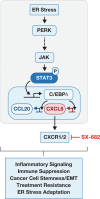
Cancer cells experience endoplasmic reticulum (ER) stress due to activated oncogenes and conditions of nutrient deprivation and hypoxia. The ensuing unfolded protein response (UPR) is executed by ATF6, IRE1 and PERK pathways. Adaptation to mild ER stress promotes tumor cell survival and aggressiveness. Unmitigated ER stress, however, will result in cell death and is a potential avenue for cancer therapies. Because of this yin-yang nature of ER stress, it is imperative that we fully understand the mechanisms and dynamics of the UPR and its contribution to the complexity of tumor biology. The PERK pathway inhibits global protein synthesis while allowing translation of specific mRNAs, such as the ATF4 transcription factor. Using thapsigargin and tunicamycin to induce acute ER stress, we identified the transcription factor C/EBPδ (CEBPD) as a mediator of PERK signaling to secretion of tumor promoting chemokines. In melanoma and breast cancer cell lines, PERK mediated early induction of C/EBPδ through ATF4-independent pathways that involved at least in part Janus kinases and the STAT3 transcription factor. Transcriptional profiling revealed that C/EBPδ contributed to 20% of thapsigargin response genes including chaperones, components of ER-associated degradation, and apoptosis inhibitors. In addition, C/EBPδ supported the expression of the chemokines CXCL8 (IL-8) and CCL20, which are known for their tumor promoting and immunosuppressive properties. With a paradigm of short-term exposure to thapsigargin, which was sufficient to trigger prolonged activation of the UPR in cancer cells, we found that conditioned media from such cells induced cytokine expression in myeloid cells. In addition, activation of the CXCL8 receptor CXCR1 during thapsigargin exposure supported subsequent sphere formation by cancer cells. Taken together, these investigations elucidated a novel mechanism of ER stress-induced transmissible signals in tumor cells that may be particularly relevant in the context of pharmacological interventions.
© 2021. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress.J Biol Chem. 2019 May 17;294(20):8197-8217. doi: 10.1074/jbc.RA118.002829. Epub 2019 Mar 29. J Biol Chem. 2019. PMID: 30926605 Free PMC article.
-
Sustained IRE1 and ATF6 signaling is important for survival of melanoma cells undergoing ER stress.Cell Signal. 2014 Feb;26(2):287-94. doi: 10.1016/j.cellsig.2013.11.008. Epub 2013 Nov 12. Cell Signal. 2014. PMID: 24240056
-
Selective, potent blockade of the IRE1 and ATF6 pathways by 4-phenylbutyric acid analogues.Br J Pharmacol. 2013 Oct;170(4):822-34. doi: 10.1111/bph.12306. Br J Pharmacol. 2013. PMID: 23869584 Free PMC article.
-
The Role of the PERK/eIF2α/ATF4/CHOP Signaling Pathway in Tumor Progression During Endoplasmic Reticulum Stress.Curr Mol Med. 2016;16(6):533-44. doi: 10.2174/1566524016666160523143937. Curr Mol Med. 2016. PMID: 27211800 Free PMC article. Review.
-
Molecular Basis of Human Diseases and Targeted Therapy Based on Small-Molecule Inhibitors of ER Stress-Induced Signaling Pathways.Curr Mol Med. 2017;17(2):118-132. doi: 10.2174/1566524017666170306122643. Curr Mol Med. 2017. PMID: 28266275 Review.
Cited by
-
Gut microbiota-derived acetic acids promoted sepsis-induced acute respiratory distress syndrome by delaying neutrophil apoptosis through FABP4.Cell Mol Life Sci. 2024 Oct 25;81(1):438. doi: 10.1007/s00018-024-05474-y. Cell Mol Life Sci. 2024. PMID: 39453486 Free PMC article.
-
Endoplasmic Reticulum Stress in Renal Cell Carcinoma.Int J Mol Sci. 2023 Mar 3;24(5):4914. doi: 10.3390/ijms24054914. Int J Mol Sci. 2023. PMID: 36902344 Free PMC article. Review.
-
Endoplasmic reticulum stress: an emerging therapeutic target for preeclampsia†.Biol Reprod. 2025 Jul 13;113(1):19-33. doi: 10.1093/biolre/ioaf071. Biol Reprod. 2025. PMID: 40156296 Free PMC article. Review.
-
Hypoxia Abrogates Tumor-Suppressive Activities of C/EBPδ in Pancreatic Cancer.Int J Mol Sci. 2024 Aug 30;25(17):9449. doi: 10.3390/ijms25179449. Int J Mol Sci. 2024. PMID: 39273396 Free PMC article.
-
The neuropathobiology of multiple sclerosis.Nat Rev Neurosci. 2024 Jul;25(7):493-513. doi: 10.1038/s41583-024-00823-z. Epub 2024 May 24. Nat Rev Neurosci. 2024. PMID: 38789516 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

