Association between prognostic factors and the outcomes of patients infected with SARS-CoV-2 harboring multiple spike protein mutations
- PMID: 34725366
- PMCID: PMC8560824
- DOI: 10.1038/s41598-021-00459-4
Association between prognostic factors and the outcomes of patients infected with SARS-CoV-2 harboring multiple spike protein mutations
Abstract
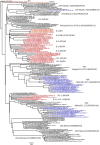
The outcome of SARS-CoV-2 infection is determined by multiple factors, including the viral, host genetics, age, and comorbidities. This study investigated the association between prognostic factors and disease outcomes of patients infected by SARS-CoV-2 with multiple S protein mutations. Fifty-one COVID-19 patients were recruited in this study. Whole-genome sequencing of 170 full-genomes of SARS-CoV-2 was conducted with the Illumina MiSeq sequencer. Most patients (47%) had mild symptoms of COVID-19 followed by moderate (19.6%), no symptoms (13.7%), severe (4%), and critical (2%). Mortality was found in 13.7% of the COVID-19 patients. There was a significant difference between the age of hospitalized patients (53.4 ± 18 years) and the age of non-hospitalized patients (34.6 ± 19) (p = 0.001). The patients' hospitalization was strongly associated with hypertension, diabetes, and anticoagulant and were strongly significant with the OR of 17 (95% CI 2-144; p = 0.001), 4.47 (95% CI 1.07-18.58; p = 0.039), and 27.97 (95% CI 1.54-507.13; p = 0.02), respectively; while the patients' mortality was significantly correlated with patients' age, anticoagulant, steroid, and diabetes, with OR of 8.44 (95% CI 1.5-47.49; p = 0.016), 46.8 (95% CI 4.63-472.77; p = 0.001), 15.75 (95% CI 2-123.86; p = 0.009), and 8.5 (95% CI 1.43-50.66; p = 0.019), respectively. This study found the clade: L (2%), GH (84.3%), GR (11.7%), and O (2%). Besides the D614G mutation, we found L5F (18.8%), V213A (18.8%), and S689R (8.3%). No significant association between multiple S protein mutations and the patients' hospitalization or mortality. Multivariate analysis revealed that hypertension and anticoagulant were the significant factors influencing the hospitalization and mortality of patients with COVID-19 with an OR of 17.06 (95% CI 2.02-144.36; p = 0.009) and 46.8 (95% CI 4.63-472.77; p = 0.001), respectively. Moreover, the multiple S protein mutations almost reached a strong association with patients' hospitalization (p = 0.07). We concluded that hypertension and anticoagulant therapy have a significant impact on COVID-19 outcomes. This study also suggests that multiple S protein mutations may impact the COVID-19 outcomes. This further emphasized the significance of monitoring SARS-CoV-2 variants through genomic surveillance, particularly those that may impact the COVID-19 outcomes.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- World Health Organization. https://www.who.int/news/item/29-06-2020-covidtimeline Accessed on July 2, 2021.
-
- World Health Organization. https://covid19.who.int/table Accessed on July 7, 2021.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

