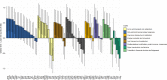Comprehensive functional core microbiome comparison in genetically obese and lean hosts under the same environment
- PMID: 34725460
- PMCID: PMC8560826
- DOI: 10.1038/s42003-021-02784-w
Comprehensive functional core microbiome comparison in genetically obese and lean hosts under the same environment
Abstract
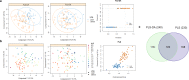
Our study provides an exhaustive comparison of the microbiome core functionalities (captured by 3,936 microbial gene abundances) between hosts with divergent genotypes for intramuscular lipid deposition. After 10 generations of divergent selection for intramuscular fat in rabbits and 4.14 phenotypic standard deviations (SD) of selection response, we applied a combination of compositional and multivariate statistical techniques to identify 122 cecum microbial genes with differential abundances between the lines (ranging from -0.75 to +0.73 SD). This work elucidates that microbial biosynthesis lipopolysaccharides, peptidoglycans, lipoproteins, mucin components, and NADH reductases, amongst others, are influenced by the host genetic determination for lipid accretion in muscle. We also differentiated between host-genetically influenced microbial mechanisms regulating lipid deposition in body or intramuscular reservoirs, with only 28 out of 122 MGs commonly contributing to both. Importantly, the results of this study are of relevant interest for the efficient development of strategies fighting obesity.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures